Reanalysis of the Cambridge reference sequence by resequencing the original placental mtDNA sample
Table from Andrews etal (1999), with permission of the authors. The full sequence of the revised CRS (rCRS) is here.
| Nucleotide Position |
1981 CRS (Anderson)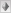 |
1999 rCRS (Andrews)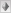 |
Remarks |
| 263 |
A |
A |
rare polymorphism |
| 311-315 |
CCCCC |
CCCCC |
rare polymorphism (5C instead of 6C) |
| 750 |
A |
A |
rare polymorphism |
| 1438 |
A |
A |
rare polymorphism |
| 3106-3107 |
CC |
C |
error |
| 3423 |
G |
T |
error |
| 4769 |
A |
A |
rare polymorphism |
| 4985 |
G |
A |
error |
| 8860 |
A |
A |
rare polymorphism |
| 9559 |
G |
C |
error |
| 11335 |
T |
C |
error |
| 13702 |
G |
C |
error |
| 14199 |
G |
T |
error |
| 14272 |
G |
C |
error (bovine) |
| 14365 |
G |
C |
error (bovine) |
| 14368 |
G |
C |
error |
| 14766 |
T |
C |
error (HeLa) |
| 15326 |
A |
A |
rare polymorphism |
"The results of the resequencing confirm that there are both errors and rare polymorphisms in the CRS. There are 11 nucleotide positions at which the original sequence contains the incorrect nucleotide (all sequences here refer to that of the CRS mtDNA L-strand), one of which involves the CC doublet at positions 3106 and 3107 in the original sequence, which is actually a single cytosine residue. The other errors are mistakes in the identification of single base pairs and typically involve the incorrect assignment of a guanine residue. Errors at nt 14272 and 14365 result from the use of the bovine mtDNA sequence at ambiguous sites. There are an additional seven nucleotide positions at which the original CRS is correct and which represent rare (or even private) polymorphic alleles. Six of these polymorphisms are single base pair substitutions, although one involves the simple repeat of cytosine residues at nt 311−315. The CRS has five residues in this repeat, whereas most human mtDNAs have six."
"The revised CRS mtDNA belongs to European haplogroup H on the basis of the cytosine at nt 14766 (instead of the thymine in the original CRS) and the cytosine at nt 7028. The assignment of the revised CRS to haplogroup H is confirmed by the absence of any of the predicted restriction site changes that characterize the other European mtDNA haplogroups."
"If we include both the technical errors (8) and the errors (3) introduced by the assumption of the bovine and
HeLa mtDNA sequences, the overall error frequency for the original CRS analysis was 0.07%, emphasizing the remarkable tour de force by Sanger and colleagues. Nevertheless, because of its widespread use, we recommend that the CRS for human mtDNA should be revised as follows: (i) the ten simple substitution errors should be corrected; (ii) the rare polymorphic alleles should be retained (that is, the revised CRS (RCRS) should be a true reference sequence and not a consensus sequence); and (iii) the original nucleotide numbering should be retained. The last suggestion represents a compromise between accuracy (correcting the numbering to account for the single C residue at nt 3106 and 3107) and consistency with the previous literature. We believe that renumbering all of the previously identified sequence changes beyond nt 3106 would create an unacceptable level of confusion."
From Andrews etal (1999), with permission of the authors  Copyright © by the contributing authors. All material on this collaboration platform is the property of the contributing authors.
Copyright © by the contributing authors. All material on this collaboration platform is the property of the contributing authors. 