Mutation Query
| | | | Allele 1: | G11D, R852C | | Allele 2: | E1143G, W748S | Allelic information known | | Refine query |
|
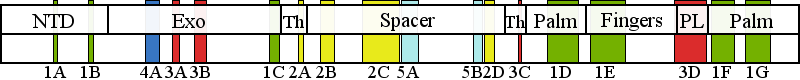
| | | Residue G11 | | Cluster assignment: | | | Cluster description: | Single Nucleotide Polymorphism | | POLG domain: | N-Terminal domain |
| Residue W748 | | Cluster assignment: | | | Cluster description: | Putative protein-protein interactions | | Subcluster: | 5B (residues 737-749) | | Subcluster description: | Located in the periphery of the IP subdomain of the spacer domain, distant from the DNA binding channel | | POLG domain: | Spacer domain |
| Residue R852 | | Cluster assignment: | | | Cluster description: | Polymerase active site and environs | | Subcluster: | 1D (residues 848-895) | | Subcluster description: | This subcluster forms a large portion of the pol active site and contains two highly conserved motifs that are found in all family A polymerases: the RR loop (motif 2) and motif A (Loh and Loeb, 2005). | | POLG domain: | Polymerase domain |
| Residue E1143 | | Cluster assignment: | | | Cluster description: | Single Nucleotide Polymorphism | | POLG domain: | Polymerase domain |
|
|
Mutation Information
|
| G11D | | | | Number of patients: (with G11D) | 11 | | Found together with: | |  | Show Patient Data |
| Patient data are sorted by mutation combination frequency. | Reference: | Ashley et al, 2008; | | Description: | reported as Alpers, onset at 2 years, presenting encephalopathy with epilepsy, hepatopathy. 22% mtDNA copy number in muscle. | | Mutations: | A467T, R852C | | SNPs: | G11D | | Age group: | infantile | | Age of Onset: 2.25, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Stewart et al, 2009; | | Description: | Alpers, seizures, abnormal liver function, mild lactic acidosis, normal histology of COX fibers. | | Mutations: | A467T, R852C | | SNPs: | G11D | | Age group: | infantile | | Age of Onset: 1.25, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Dementia/encephalopathy, infantile spasms, cerebral artery occlusion, headache/migraine, stroke, constipation. 74% mtDNA copy number in blood. | | Mutations: | R627Q, R852C | | SNPs: | G11D | | Age group: | juvenile | | Age of Onset: n/a, Age of Patient: 25, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Seizures, myoclonic seizures, intractable seizures, hepatic failure, elevated transaminases, Apnes/Hypoventilation, respiratory deficiency/failure, lactic acidosis, organic aciduria/tiglyglycine, low plasma carnitine, abnormal EEG. 108% mtDNA copy number in blood. | | Mutations: | A467T, R852C | | SNPs: | G11D | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 0.8, Age of Death: n/a |
Back to top | Reference: | Wong et al, 2008; | | Description: | Ataxia Neuropathy Spectrum, Seizures, PEO, lactic acidosis, stroke, chorea, ataxia, ptosis, retinitis pigmentosa, liver transplans | | Mutations: | R627Q, R852C | | SNPs: | G11D | | Age group: | juvenile | | Age of Onset: 15, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Seizures, PEO, Ataxia Neuropathy Spectrum, Stroke, chorea, ataxia, ptosis, retinitis pigmentosa, liver transplant | | Mutations: | R627Q, R852C | | SNPs: | G11D | | Age group: | juvenile | | Age of Onset: 15, Age of Patient: 19, Age of Death: n/a |
| Reference: | Mehta et al, 2011; | | Description: | progressive cerebellar syndrome, slurred speech, balance disturbances, mild incoordination, type II diabetes mellitus, dysarthria, mild limb dysmetria, mildly impaired tandem gait, cerebellar ataxia. She had polyminimyoclonus of her outstretched hands, a positive glabellar tap and brisk deep tendon reflexes with flexor plantar responses. She had Rigidity in the legs worse than in the arms, brisk reflexes, and mild bilateral bradykinesia in association with dystonic posturing of the left hand. Dystonia. | | Mutations: | R852C | | SNPs: | G11D | | Age group: | adult | | Age of Onset: 49, Age of Patient: 58, Age of Death: n/a |
| Reference: | Ashley et al, 2008; | | Description: | reported as Alpers, onset at 1 years, presenting encephalopathy with epilepsy, hepatopathy, and movement disorder (ataxia). 32% mtDNA copy number in liver. | | Mutations: | R852C, W748S | | SNPs: | G11D | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Stewart et al, 2009; | | Description: | Alpers, Seizures, Epilepsia partialis continua, liver failure, stroke-like episode. | | Mutations: | R852C, W748S | | SNPs: | E1143G, G11D | | Age group: | infantile | | Age of Onset: 1.33, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Naess et al, 2009; | | Description: | Developmental retardation, Hypotonia, ataxia, seizures myoclonus, abnormal eye movement, gastrointestinal problems, hepatic involvement | | Mutations: | R852C, W748S | | SNPs: | G11D | | Age group: | infantile | | Age of Onset: 0.3, Age of Patient: n/a, Age of Death: 1.1 |
| Reference: | Vasta et al, 2012; | | Description: | PEO, alpers disease, ataxia, seizure, hypotonia, developemental delay, | | Mutations: | R852C, W748S | | SNPs: | E1143G, G11D | | Age group: | infantile | | Age of Onset: 0.001, Age of Patient: n/a, Age of Death: n/a |
|
| R852C | | | | Number of patients: (with R852C) | 15 | | Found together with: | |  | Show Patient Data |
| Patient data are sorted by mutation combination frequency. | Reference: | Ashley et al, 2008; | | Description: | reported as Alpers, onset at 2 years, presenting encephalopathy with epilepsy, hepatopathy. 22% mtDNA copy number in muscle. | | Mutations: | A467T, R852C | | SNPs: | G11D | | Age group: | infantile | | Age of Onset: 2.25, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Stewart et al, 2009; | | Description: | Alpers, seizures, abnormal liver function, mild lactic acidosis, normal histology of COX fibers. | | Mutations: | A467T, R852C | | SNPs: | G11D | | Age group: | infantile | | Age of Onset: 1.25, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Seizures, myoclonic seizures, intractable seizures, hepatic failure, elevated transaminases, Apnes/Hypoventilation, respiratory deficiency/failure, lactic acidosis, organic aciduria/tiglyglycine, low plasma carnitine, abnormal EEG. 108% mtDNA copy number in blood. | | Mutations: | A467T, R852C | | SNPs: | G11D | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 0.8, Age of Death: n/a |
| Reference: | Mehta et al, 2011; | | Description: | progressive cerebellar syndrome, slurred speech, balance disturbances, mild incoordination, type II diabetes mellitus, dysarthria, mild limb dysmetria, mildly impaired tandem gait, cerebellar ataxia. She had polyminimyoclonus of her outstretched hands, a positive glabellar tap and brisk deep tendon reflexes with flexor plantar responses. She had Rigidity in the legs worse than in the arms, brisk reflexes, and mild bilateral bradykinesia in association with dystonic posturing of the left hand. Dystonia. | | Mutations: | R852C | | SNPs: | G11D | | Age group: | adult | | Age of Onset: 49, Age of Patient: 58, Age of Death: n/a |
Back to top | Reference: | Hunter et al, 2011; | | Description: | Myoclonic Seizures, visual disturbance, motor paresis, Focal motor seizures, Epilepsia partialis continua, Developmental Delay or Regression, ataxia, Serum Lactate, liver mtDNA depletion, Alpers | | Mutations: | A467T, R852C | | Age group: | infantile | | Age of Onset: 1.33, Age of Patient: n/a, Age of Death: 5.08 |
| Reference: | McCoy et al, 2011; | | Description: | clonic seizures of the right arm and leg, Coma persisted and was accompanied by hypoglycaemia, tachycardia and oliguria, CSF proteinwas elevated, cortical blindness, anarthria and profound hypotonia, Liver dysfunction, prolonged focal seizures, acute encephalopathy, | | Mutations: | A467T, R852C | | Age group: | infantile | | Age of Onset: 1.75, Age of Patient: n/a, Age of Death: 3.5 |
| Reference: | Ashley et al, 2008; | | Description: | reported as Alpers, onset at 1 years, presenting encephalopathy with epilepsy, hepatopathy, and movement disorder (ataxia). 32% mtDNA copy number in liver. | | Mutations: | R852C, W748S | | SNPs: | G11D | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Stewart et al, 2009; | | Description: | Alpers, Seizures, Epilepsia partialis continua, liver failure, stroke-like episode. | | Mutations: | R852C, W748S | | SNPs: | E1143G, G11D | | Age group: | infantile | | Age of Onset: 1.33, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Naess et al, 2009; | | Description: | Developmental retardation, Hypotonia, ataxia, seizures myoclonus, abnormal eye movement, gastrointestinal problems, hepatic involvement | | Mutations: | R852C, W748S | | SNPs: | G11D | | Age group: | infantile | | Age of Onset: 0.3, Age of Patient: n/a, Age of Death: 1.1 |
| Reference: | Vasta et al, 2012; | | Description: | PEO, alpers disease, ataxia, seizure, hypotonia, developemental delay, | | Mutations: | R852C, W748S | | SNPs: | E1143G, G11D | | Age group: | infantile | | Age of Onset: 0.001, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Sofou et al, 2013; | | Description: | Alpers/ Alpers–Huttenlocher. | | Mutations: | Q497H, R852C, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Dementia/encephalopathy, infantile spasms, cerebral artery occlusion, headache/migraine, stroke, constipation. 74% mtDNA copy number in blood. | | Mutations: | R627Q, R852C | | SNPs: | G11D | | Age group: | juvenile | | Age of Onset: n/a, Age of Patient: 25, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Ataxia Neuropathy Spectrum, Seizures, PEO, lactic acidosis, stroke, chorea, ataxia, ptosis, retinitis pigmentosa, liver transplans | | Mutations: | R627Q, R852C | | SNPs: | G11D | | Age group: | juvenile | | Age of Onset: 15, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Wong et al, 2008; | | Description: | Seizures, PEO, Ataxia Neuropathy Spectrum, Stroke, chorea, ataxia, ptosis, retinitis pigmentosa, liver transplant | | Mutations: | R627Q, R852C | | SNPs: | G11D | | Age group: | juvenile | | Age of Onset: 15, Age of Patient: 19, Age of Death: n/a |
|
| E1143G | | | | Number of patients: (with E1143G) | 95 | | Found as the only mutation: | 1% of entries (1 patient) | | Found together with: | PNF=Putatively Non-Functional enzyme |  | Show Patient Data |
| Patient data are sorted by mutation combination frequency. | Reference: | Blok et al, 2009; | | Description: | Ptosis, CPEO, polyneuropathy, cerebellar ataxia, dysarthria. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 47, Age of Death: n/a |
| Reference: | Ferrari et al, 2005; | | Description: | An apparently myogenic torticollis was noticed after a few months of life; at 10 months, he had two episodes of sudden head drop, followed by the onset of psychomotor arrest/ regression, hypotonia, ataxia and focal myoclonus of the right upper limb. In the following weeks, the myoclonus became subcontinuous, and was associated with rapidly progressive, severe generalized hypotonia and weakness requiring ventilatory assistance. Seizures were partially controlled with a combination of topiramate, clobazam and phenylbarbiturate. symmetrical lesions of the basal ganglia, thalami, cerebellar dentate nuclei and left occipital cortical and subcortical regions. abnormal accumulation of lactic acid in the putamen and a reduction of the N-acetyl aspartate. cholestatic jaundice, hypoglycaemia and hypocoagulation. The diagnostic conclusion was Alpers syndrome. | | Mutations: | W748S | | SNPs: | E1143G, PNF | | Age group: | infantile | | Age of Onset: 0.2, Age of Patient: n/a, Age of Death: 2.5 |
| Reference: | Ferrari et al, 2005; | | Description: | disease-free interval in the first 4 months after birth, followed by persistent vomiting, epileptic crises with loss of eye contact, and myoclonus in the upper limbs with secondary generalization and loss of consciousness. Persistent myoclonus in the upper limbs was associated with profound hypotonia and loss of eye contact. cortical atrophy and leukoencephalopathy in the subcortical areas of the frontal lobes. progressive hepatic failure with enlarged liver, hypoalbuminaemia and predominantly cholestatic jaundice led to the death of the child at 8 months of age. | | Mutations: | W748S | | SNPs: | E1143G, PNF | | Age group: | infantile | | Age of Onset: 0.3, Age of Patient: n/a, Age of Death: 0.67 |
| Reference: | Isohanni et al, 2011; | | Description: | Partial status epilepticus, epilepsia partialis, continua, hemiparesis, myoclonus, loss of vision, optical atrophy, pschomotor regression, liver dysfunction, death via pneumonia. | | Mutations: | G848S, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: 11 |
Back to top | Reference: | Isohanni et al, 2011; | | Description: | developmental delay, vomiting, partial status epilepticus, epilepsia partialis continua, myoclonus, ataxia, alive at age 3. | | Mutations: | T914P, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 2.5, Age of Patient: 3, Age of Death: n/a |
| Reference: | Isohanni et al, 2011; | | Description: | epilepsy, vomiting, lowered consciousness, partial status epilepticus, epilepsia partialis continua, myoclonus, pschomotor regression, liver failure before valproate treatment, death via pneumonia. 72-76% mtDNA copy number in muscle. | | Mutations: | R807C, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: 3 |
| Reference: | Kollberg et al, 2006; | | Description: | Alpers, age of onset 6 months, age of death 13 months. 41% mtDNA copy number. During the first 6 months of life, he was considered healthy, but was irritable with colic-like symptoms. failure to thrive, delayed motor development. myoclonus. leftsided hemiparesis. | | Mutations: | R232H, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: n/a, Age of Death: 1.08 |
| Reference: | Kollberg et al, 2006; | | Description: | Ragged Red Fibers, COX-Deficient Fibers. mtDNA depletion. He was healthy and his development was normal until 5 months of age. He developed failure to thrive and muscular hypotonia. At 4 years of age, he developed myoclonus, which was first limited to the right eye and the right side of the mouth, but then progressed to epilepsia partialis continua of the entire left side of the body without loss of consciousness. He has developed a complex movement disorder and cognitive impairment, but contact, speech, and memory functions have been retained. He also had ptosis and uncontrolled and uncoordinated movements, especially in his arms. | | Mutations: | E1163G, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 0.417, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Kollberg et al, 2006; | | Description: | Started as an ataxic syndrome, Alpers, ragged red fibers, COX-deficient fibers, mtDNA deletions. mtDNA copy numbers 1.6 fold. She was healthy and developed normally until her late preschool years when she developed slowly progressive ataxia. Occasional myoclonic seizures, and at 13 years of age, valproate treatment was started because of occipital seizures. ataxia, dementia, and cortical blindness. She developed refractory seizures, including multifocal EPC, status epilepticus, and myoclonus. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | childhood | | Age of Onset: 5, Age of Patient: 13, Age of Death: 14 |
Back to top | Reference: | Kollberg et al, 2006; | | Description: | psychomotor regression, refractory seizures, stroke-like episodes, hepatopathy, and ataxia. COX-deficient muscle fibers, mtDNA deletions, 160% mtDNA copy number. He was normal until 11 years of age when he developed mild tremor and ataxia. At 13 years of age, he reported occasional migraine-like headaches and was found to have mild ataxia and myoclonus. generalized seizures. mild leftsided hemiparesis, external ophthalmoplegia, and peripheral neuropathy. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | childhood | | Age of Onset: 11, Age of Patient: 24, Age of Death: n/a |
| Reference: | Naimi et al, 2006; | | Description: | SCAE, age of onset 20, death at 27, ptosis and multiple deletions in muscle. | | Mutations: | L304R, W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: 20, Age of Patient: n/a, Age of Death: 27 |
| Reference: | Sarzi et al, 2007; | | Description: | Myoclonus, epilepsy, Psychomotor retardation, Alpers syndrome, steatosis | | Mutations: | W748S | | SNPs: | E1143G, PNF | | Age group: | infantile | | Age of Onset: 0.25, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Sarzi et al, 2007; | | Description: | Onset with walking difficulties, seizures at 2.5 years, hepatocellular insufficiency after valproate treatment, alpers syndrome, Liver mtDNA depletion. | | Mutations: | W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 2, Age of Patient: n/a, Age of Death: 3 |
| Reference: | Stewart et al, 2009; | | Description: | Alpers, Seizures, Epilepsia partialis continua, liver failure, stroke-like episode. | | Mutations: | R852C, W748S | | SNPs: | E1143G, G11D | | Age group: | infantile | | Age of Onset: 1.33, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Taanman et al, 2009; | | Description: | Hypotonia at 8 weeks, seizures, hepatopathy, lactic acidemia, died of liver failure. 9% mtDNA copy number in muscle, 11% mtDNA copy number in liver. | | Mutations: | H1110Y, W748S | | SNPs: | E1143G, Q1236H | | Age group: | infantile | | Age of Onset: 0.1, Age of Patient: n/a, Age of Death: 0.8 |
| Reference: | Taanman et al, 2009; | | Description: | Onset at 6.5 years with alpers syndrome and death at 7.8 years. 23% mtDNA copy number in liver. | | Mutations: | G848S, W748S | | SNPs: | E1143G | | Age group: | childhood | | Age of Onset: 6.5, Age of Patient: n/a, Age of Death: 7.8 |
| Reference: | Winterthun et al, 2005; | | Description: | Ataxia, sensory ataxia, dysarthria, nystagmus, cognitive dysfunction, axonal neuropathy, demyelinating neuropathy. This man presented at age 26 with unsteadiness. Examination showed an ataxic gait, cerebellar dysarthria, myoclonic jerks of his head and facial muscles, mild limitation of horizontal eye movement, and horizontal nystagmus in the direction of gaze, with an additional vertical element when looking down. There were distal amyotrophy, absent reflexes in the legs, and a symmetric loss of all sensory modalities below the knee. Romberg’s test was positive. Episodes of depression. At age 31 he has an almost complete ophthalmoplegia, cerebellar dysarthria, myoclonus involving face and arms, truncal ataxia, and symmetric dysmetria. | | Mutations: | Q497H, Q497H, W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 23, Age of Patient: 38, Age of Death: n/a |
| Reference: | Winterthun et al, 2005; | | Description: | Headaches, a focal epilepsy with secondary generalisation, occipital epilepsy, dysarthria, nystagmus, cognitive dysfunction. Demyelinating neuropathy, axonal neuropathy. headaches preceded by visual symptoms, nausea, vomiting, and unsteadiness diagnosed as migraine. Shortly after, she had the first of a series of tonic-clonic seizures preceded by headache. Examination showed horizontal and vertical nystagmus, gait ataxia. | | Mutations: | Q497H, Q497H, W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | juvenile | | Age of Onset: 15, Age of Patient: 18, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Hearing loss, Ataxia Neuropathy Spectrum, Seizures, dementia, developmental delay | | Mutations: | A467T, Q497H, W748S | | SNPs: | E1143G | | Age group: | juvenile | | Age of Onset: 17, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Wong et al, 2008; | | Description: | Muscle weakness, respiratory failure, Alpers, Lactic acidosis, Seizures, Demential , developmental delay | | Mutations: | W748S | | SNPs: | E1143G, PNF | | Age group: | childhood | | Age of Onset: 4, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Onset 33 years with neuropathy, myopathy, SANDO, lactic acidosis, PEO. | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 33, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | developmental delay, dementia, seizures, Alpers, Family history of Alpers syndrome. | | Mutations: | G848S, Q497H, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Zsurka et al, 2008; | | Description: | Epilepsy, seizures, VPA induced liver failure. | | Mutations: | L752P, W748S | | SNPs: | E1143G | | Age group: | childhood | | Age of Onset: 7, Age of Patient: n/a, Age of Death: 10 |
| Reference: | Davidzon et al, 2005; | | Description: | hepatocerebral syndrome, alpers, developmental delay, seizures, and hepatopathy leading to liver failure, liver mtDNA depletion | | Mutations: | G848S, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 0.7, Age of Patient: n/a, Age of Death: 2.5 |
Back to top | Reference: | Davidzon et al, 2005; | | Description: | hepatocerebral syndrome, alpers, developmental delay, seizures, and hepatopathy leading to liver failure, liver mtDNA depletion, microvesicular steatosis, | | Mutations: | G848S, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: 6.5 |
| Reference: | Davidzon et al, 2005; | | Description: | hepatocerebral syndrome, alpers, developmental delay, seizures, and hepatopathy leading to liver failure, liver mtDNA depletion, microvesicular steatosis. | | Mutations: | G848S, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 0.1, Age of Patient: n/a, Age of Death: 0.9 |
| Reference: | Davidzon et al, 2005; | | Description: | hepatocerebral syndrome, alpers, developmental delay, seizures, and hepatopathy leading to liver failure, microvesicular steatosis, fibrosis | | Mutations: | G848S, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 0.3, Age of Patient: n/a, Age of Death: 1 |
| Reference: | Nguyen et al, 2005; | | Description: | Refractory seizures, psychomotor regression, liver disease, presenting symptom was status epilepticus, Transient hypoglycemia, Transient lactic acidemia | | Mutations: | G848S, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: 1.7 |
| Reference: | Milone et al, 2011; | | Description: | Stiffness, cramping of the lower extremities, foot numbness, and poor balance. The past medical history was significant for hypogonadotropic hypogonadism diagnosed 10 years prior. ophthalmoplegia without ptosis, mild lower proximal weakness, Multiple mtDNA deletions detected by PCR in muscle | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 32, Age of Patient: 33, Age of Death: n/a |
Back to top | Reference: | Milone et al, 2011; | | Description: | progressive unsteadiness,limb paresthesias, dysarthria, dysphagia, and weakness, ophthalmoparesis,dysarthria, palatal tremor, moderate axial and appendicular ataxia, and distal pan-modality sensory loss | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 32, Age of Patient: 33, Age of Death: n/a |
| Reference: | Komulainen et al, 2010; | | Description: | mental retardation, psychiatric symptoms, mild bilateral ptosis and epilepsy, Seizures occurred at age 11 years, and focal generalized epilepsy was diagnosed. | | Mutations: | W748S | | SNPs: | E1143G, R722H | | Age group: | childhood | | Age of Onset: 11, Age of Patient: 22, Age of Death: n/a |
| Reference: | Mousson de Camaret et al, 2011 | | Description: | At 16 months of age, he started a severe status epilepticus. Cerebral MRI showed brain cortical atrophy. he developed intractable epileptic encephalopathy associated with generalized hypotonia and cortical blindness. diffuse hypotonia, lethargy and vomiting. During the two following years, epilepsy was always very active including spasms and myoclonic jerks. progressive hepatic failure. Alpers. | | Mutations: | W748S | | SNPs: | E1143G, PNF | | Age group: | infantile | | Age of Onset: 1.33, Age of Patient: n/a, Age of Death: 4.17 |
| Reference: | Vasta et al, 2012; | | Description: | PEO, alpers disease, ataxia, seizure, hypotonia, developemental delay, | | Mutations: | R852C, W748S | | SNPs: | E1143G, G11D | | Age group: | infantile | | Age of Onset: 0.001, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Hakonen et al, 2005; | | Description: | Onset with balance disturbances and neuropathy, Gait and limb ataxia, Dysarthria, restricted eye movements, ptosis, Cognitive impairment, Obesity, cramps, sensory motor polyneuropathy | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 38, Age of Patient: 44, Age of Death: n/a |
Back to top | Reference: | Hakonen et al, 2005; | | Description: | Onset with balance disturbances and neuropathy, Gait and limb ataxia, Myoclonus, tremor, Dysarthria, restricted eye movements, Cognitive impairment, Psychiatric symptoms, Obesity, sensory motor polyneuropathy | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 38, Age of Patient: 51, Age of Death: n/a |
| Reference: | Hakonen et al, 2005; | | Description: | Onset with balance disturbances and neuropathy, Gait and limb ataxia, tremor, Involuntary movements, Dysarthria, Dysphagia, Nystagmus, restricted eye movements, Psychiatric symptoms, Muscle strength decreased, Obesity, muscle cramps, amyotrophy, pes cavus, sensory motor polyneuropathy | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 27, Age of Patient: 38, Age of Death: n/a |
| Reference: | Hakonen et al, 2005; | | Description: | Onset with balance disturbances, Gait and limb ataxia, Dysarthria, Dysphagia, Nystagmus, diplopia, Cognitive impairment, Muscle strength decreased, sensory neural hearing deficit, sensory motor polyneuropathy | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 32, Age of Patient: 46, Age of Death: n/a |
| Reference: | Hakonen et al, 2005; | | Description: | Onset with balance disturbances, Gait and limb ataxia, facial Involuntary movements, Dysarthria, Dysphagia, Nystagmus, other eye-movement abnormalities, Psychiatric symptoms, premature menopause, Obesity, sensory motor polyneuropathy | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 27, Age of Patient: 42, Age of Death: n/a |
| Reference: | Hakonen et al, 2005; | | Description: | Onset with epilepsy, Gait and limb ataxia, Epilepsy, Dysarthria, Nystagmus, diplopia, Psychiatric symptoms, Obesity, sensory motor polyneuropathy | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 24, Age of Patient: 55, Age of Death: n/a |
Back to top | Reference: | Hakonen et al, 2005; | | Description: | Onset with epilepsy, Gait and limb ataxia, Epilepsy, Myoclonus, facial Involuntary movements, Dysarthria, Dysphagia, Nystagmus, other eye-movement abnormalities, Cognitive impairment, Psychiatric symptoms, Muscle strength decreased, Obesity | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | juvenile | | Age of Onset: 16, Age of Patient: 37, Age of Death: n/a |
| Reference: | Hakonen et al, 2005; | | Description: | Onset with epilepsy, Gait and limb ataxia, Epilepsy, Myoclonus, head tremor, Dysarthria, other eye-movement abnormalities, Nystagmus, Cognitive impairment, Psychiatric symptoms, cramps, sensory motor polyneuropathy | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 23, Age of Patient: 45, Age of Death: n/a |
| Reference: | Hakonen et al, 2005; | | Description: | Onset with epilepsy, Gait and limb ataxia, Epilepsy, Myoclonus, head tremor, facial Involuntary movements, Dysarthria, Nystagmus, other eye-movement abnormalities, Cognitive impairment, Psychiatric symptoms, cramps | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | juvenile | | Age of Onset: 17, Age of Patient: 45, Age of Death: n/a |
| Reference: | Hakonen et al, 2005; | | Description: | Onset with epilepsy, Gait and limb ataxia, Myoclonus, Involuntary movements, Dysarthria, Dysphagia, Nystagmus, other eye-movement abnormalities, Cognitive impairment, Obesity, sensory neural hearing deficit, sensory motor polyneuropathy | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | juvenile | | Age of Onset: 19, Age of Patient: 44, Age of Death: n/a |
| Reference: | Hakonen et al, 2005; | | Description: | Onset with headaches, Gait and limb ataxia, Epilepsy, Myoclonus, Involuntary movements, Dysarthria, Dysphagia, Nystagmus, other eye-movement abnormalities, restricted eye movements, ptosis, Cognitive impairment, Psychiatric symptoms, Muscle strength decreased, Obesity | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 28, Age of Patient: 51, Age of Death: n/a |
Back to top | Reference: | Hakonen et al, 2005; | | Description: | Onset with neuropathy, Gait and limb ataxia, Dysarthria, Dysphagia, other eye-movement abnormalities, Cognitive impairment, Psychiatric symptoms, Obesity, sensory motor polyneuropathy | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 41, Age of Patient: 58, Age of Death: n/a |
| Reference: | Hakonen et al, 2005; | | Description: | Onset with neuropathy, Gait and limb ataxia, tremor, Dysarthria, Nystagmus, Psychiatric symptoms, Muscle strength decreased, amyotrophy, pes cavus, sensory motor polyneuropathy | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 36, Age of Patient: 51, Age of Death: n/a |
| Reference: | Hakonen et al, 2005; | | Description: | Onset with tremor, Gait and limb ataxia, Epilepsy, Myoclonus, Involuntary movements, Dysarthria, eye muscle weakness, Cognitive impairment, Psychiatric symptoms, Muscle strength decreased. | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | childhood | | Age of Onset: 5, Age of Patient: n/a, Age of Death: 35 |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with epilepsy and headaches, ataxia, status epilepticus, nystagmus, myoclonus, neuropathy. Sodium valproate treatment 6 weeks prior to death. Liver dysfunction, cause of death status epilepticus and liver failure. Hepatic histology showed acute liver necrosis. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | juvenile | | Age of Onset: 19, Age of Patient: n/a, Age of Death: 19 |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with epilepsy and headaches, ataxia, epilepsy, status epilepticus, myoclonus, neuropathy. Sodium valproate treatment 7 weeks prior to death. Liver dysfunction and rising liver enzymes and bilirubin terminally, cause of death status epilepticus. Hepatic histology showed centrolobular degeneration. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | juvenile | | Age of Onset: 14, Age of Patient: n/a, Age of Death: 23 |
Back to top | Reference: | Tzoulis et al, 2006; | | Description: | Presented with epilepsy, ataxia, nystagmus, neuropathy, ptosis/ PEO onset at age 65 | | Mutations: | W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: 55, Age of Patient: 74, Age of Death: n/a |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with epilepsy, ataxia, status epilepticus, headaches, myoclonus, neuropathy | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | juvenile | | Age of Onset: 18, Age of Patient: 19, Age of Death: n/a |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with epilepsy, ataxia, status epilepticus, headaches, myoclonus, neuropathy, ptosis/ PEO onset at age 28. Acute liver failure after 4 months of sodium valproate treatment | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | juvenile | | Age of Onset: 19, Age of Patient: 33, Age of Death: n/a |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with epilepsy, ataxia, status epilepticus, headaches, myoclonus, neuropathy. Sodium valproate treatment for 8 months. Stopped just prior to death. Liver dysfunction, cause of death liver failure. Hepatic histology showed acute liver necrosis. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | childhood | | Age of Onset: 10, Age of Patient: n/a, Age of Death: 10 |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with epilepsy, ataxia, status epilepticus, headaches, nystagmus, myoclonus, neuropathy, treatment with sodium valproate for 2 months, died 3 years later due to status epilepticus, disseminated intravascular coagulation, liver failure. | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | childhood | | Age of Onset: 10, Age of Patient: n/a, Age of Death: 22 |
Back to top | Reference: | Tzoulis et al, 2006; | | Description: | Presented with epilepsy, headaches, epilepsy, status epilepticus, headaches, nystagmus, and neuropathy. Sodium valproate treatment for 2 months then stopped 2 months prior to death. Liver dysfunction, liver transplant 1 month prior to death, cause of death liver failure. Hepatic histology showed acute liver necrosis. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: 20, Age of Patient: n/a, Age of Death: 20 |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with epilepsy, status epilepticus, headaches, nystagmus, myoclonus, neuropathy. Sodium valproate treatment 7 weeks prior to death. Liver dysfunction and rising liver enzymes and bilirubin terminally, cause of death status epilepticus. Hepatic histology showed centrolobular degeneration. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | juvenile | | Age of Onset: 15, Age of Patient: n/a, Age of Death: 21 |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with epilepsy, status epilepticus, nystagmus, neuropathy | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | juvenile | | Age of Onset: 17, Age of Patient: 18, Age of Death: n/a |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with migraine like headaches, ataxia, epilepsy, status epilepticus, nystagmus, myoclonus, neuropathy, ptosis/ PEO onset at age 26 | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | juvenile | | Age of Onset: 17, Age of Patient: 38, Age of Death: n/a |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with migraine like headaches, ataxia, epilepsy, status epilepticus, nystagmus, myoclonus, neuropathy. Sodium valproate treatment 3 months prior to death. Liver dysfunction and rising liver enzymes and bilirubin terminally, cause of death status epilepticus and multi-organ failure. Acute liver necrosis | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | childhood | | Age of Onset: 8, Age of Patient: n/a, Age of Death: 9 |
Back to top | Reference: | Tzoulis et al, 2006; | | Description: | Presented with migraine like headaches, ataxia, epilepsy, status epilepticus, nystagmus, neuropathy | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | juvenile | | Age of Onset: 17, Age of Patient: 27, Age of Death: n/a |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with migraine like headaches, ataxia, nystagmus, neuropathy, PEO/ Ptosis onset age >30 | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | childhood | | Age of Onset: 12, Age of Patient: 38, Age of Death: n/a |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with progressive gait unsteadiness and headaches, ataxia, epilepsy, status epilepticus, nystagmus, neuropathy. Died 2 months after treatment with sodium valproate, cause of death status epilepticus | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | juvenile | | Age of Onset: 15, Age of Patient: n/a, Age of Death: 57 |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with progressive gait unsteadiness, ataxia, epilepsy, status epilepticus, headaches, myoclonus, neuropathy, ptosis/ PEO onset age 30. Mild liver dysfunction. Cause of death status epilepticus. | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | childhood | | Age of Onset: 4, Age of Patient: n/a, Age of Death: 30 |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with progressive gait unsteadiness, ataxia, headaches, myoclonus, neuropathy, PEO, ptosis onset at age 44 | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: 36, Age of Patient: 50, Age of Death: n/a |
Back to top | Reference: | Tzoulis et al, 2006; | | Description: | Presented with progressive gait unsteadiness, ataxia, headaches, neuropathy, PEO ptosis onset age 25 | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: 24, Age of Patient: 43, Age of Death: n/a |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with speech delay, ataxia, epilepsy, status epilepticus, and headaches. Liver dysfunction and rising liver enzymes and bilirubin terminally, cause of death status epilepticus, multi-organ failure. Hepatic histology showed marked steatosis. | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | childhood | | Age of Onset: 8, Age of Patient: n/a, Age of Death: 9 |
| Reference: | Paus et al, 2008; | | Description: | Presented with PEO, mild bilateral ptosis, dysarthria, ataxia, and neuropathy | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: 36, Age of Patient: 37, Age of Death: n/a |
| Reference: | Remes et al, 2008; | | Description: | patient noticed gait and balance difficulties at age 46 years, ataxia, CPEO, cortical and cerebellar atrophy, axonal sensory polyneuropathy, Symmetrical bradykinesia and rigidity, mild tremor, paranoid delusions, During the last 2 years of life he had marked rigidity, dysphagia, myoclonic jerks and dystonia | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 46, Age of Patient: 65, Age of Death: n/a |
| Reference: | Uusimaa et al, 2008; | | Description: | Headache, visual symptoms, migraine-like headache, Athetosis, nystagmus, emiparesis, valproic acid induced Liver failure, sepsis, pancreatitis, status epilepticus | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | juvenile | | Age of Onset: 14, Age of Patient: 15, Age of Death: n/a |
Back to top | Reference: | Uusimaa et al, 2008; | | Description: | Migraine like headaches, visual symptoms, focal generalized seizures, severe liver failure, valproate induced liver failure | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | juvenile | | Age of Onset: 15, Age of Patient: n/a, Age of Death: 20 |
| Reference: | Uusimaa et al, 2008; | | Description: | Seizures, status epilepticus, visual symptoms, nystagmus, valproic acid induced liver failure | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | juvenile | | Age of Onset: 17, Age of Patient: 21, Age of Death: n/a |
| Reference: | Amiot et al, 2009; | | Description: | Peripheral neuropathy, PEO, ataxia, hearing loss, dysarthria-dysphonia, urinary tract involvement | | Mutations: | A143V, W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: 40, Age of Patient: 49, Age of Death: n/a |
| Reference: | Palin et al, 2012; | | Description: | Gait disturbance since childhood. Early onset suggested anticipation. In his 20's he developed photophobia and general clumsiness and benign paroxysmal positional vertigo. From the age of 37 he has had unspecific sensory polyneuropathy, confirmed by electromyography. He had several simple partial seizures at the age of 39, and has mild anxiety and depression. | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | childhood | | Age of Onset: 5, Age of Patient: 41, Age of Death: n/a |
| Reference: | Palin et al, 2012; | | Description: | No neurological symptoms, but type-2 diabetes and hypertension. | | Mutations: | W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 63, Age of Death: n/a |
Back to top | Reference: | Palin et al, 2012; | | Description: | developed gait disturbance by the age of 35, progressing to increasing clumsiness in lower extremities, dysarthria, diplopia, and occasional amnesia at the age of 44. She developed ataxia, slight polyneuropathy, and external ophthalmoplegia. At the age of 46 she had slightly increased plasma creatine kinase levels and symmetrical cerebellar peduncular white matter signal intensity increase in brain MRI. The first epileptic seizure, requiring treatment by general anesthesia, oc- curred at the age of 55, after which she was hospitalized permanently. From her 30's, she received psychiatric care due to anxiety and depression. A neuropsychological examination revealed decrease in visual reasoning and memory functions. She deceased at the age of 56 due to pneumonia and pulmonary embolism. | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 35, Age of Patient: n/a, Age of Death: 56 |
| Reference: | Posada et al, 2010; | | Description: | Ataxic sensory neuropathy, dysarthria, progressive ophthalmoplegia, paresthesia of lower extremities, unsteady gait, slurred speech, absent deep tendon reflexes, vibrations, ataxic gait and positive Romberg sign. Multiple mtDNA deletions, a marked decrease in mtDNA copy number. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: 37, Age of Patient: 47, Age of Death: n/a |
| Reference: | Sofou et al, 2013; | | Description: | Alpers/ Alpers–Huttenlocher. | | Mutations: | Q497H, R852C, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Brunetti-Pierri et al, 2008; | | Description: | He exhibited a very similar phenotype to his sister, with a catastrophic onset of intractable seizures at the age of 9 months, progressive encephalopathy with epilepsia partialis continua and myoclonic jerks, and developmental regression. MRI imaging was not performed. | | Mutations: | G848S, Q497H, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: 1.25, Age of Death: n/a |
| Reference: | Brunetti-Pierri et al, 2008; | | Description: | He exhibited a very similar phenotype to his sister, with a catastrophic onset of intractable seizures at the age of 9 months, progressive encephalopathy with epilepsia partialis continua and myoclonic jerks, and developmental regression. MRI imaging was not performed. | | Mutations: | G848S, Q497H, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: 1.25, Age of Death: n/a |
Back to top | Reference: | Brunetti-Pierri et al, 2008; | | Description: | The index patient presented at the age of 8 months with microcephaly, seizures, and developmental delay. From the time of the first observation she rapidly developed epilepsia partialis continua, multifocal myoclonic jerks, and progressive psychomotor retardation. MRI performed at 8 months was generally unremarkable. By 9 months of age the brain MRI showed signifi cant changes including a diffuse severe cerebral and cerebellar atrophy involving gray and white matter. | | Mutations: | G848S, Q497H, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 0.6, Age of Death: 1 |
| Reference: | Ylönen et al, 2013; | | Description: | late-onset parkinsons. Tremor. Cardiac arrhythmia. | | Mutations: | W748S | | SNPs: | E1143G, R993C | | Age group: | adult | | Age of Onset: 72, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Van Goethem et al, 2004; | | Description: | minor tremor of the hands from age 12 years. In his late 20s, he experienced tingling in both hands, followed by tingling in the legs. In the early fourth decade, he became unsteady at walking, especially in the dark. From his early 40s, his speech became slurred (dysarthria). On examination at age 52 years, he had an ataxic gait, and Romberg test was positive. There was dysarthria and bilateral ophthalmoparesis. generalized areflexia. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | childhood | | Age of Onset: 12, Age of Patient: 52, Age of Death: n/a |
| Reference: | Van Goethem et al, 2004; | | Description: | plurimetabolic syndrome. At age 30 years, he noted disturbed balance, and ataxia and severe axonal neuropathy were diagnosed. On examination at age 32 years, he had slowed ocular pursuit movements but with full range of motion. He had severe gait ataxia, imbalance, and trunk ataxia, as well as clumsiness of the hands. Achilles tendon reflexes were absent. dysarthria, obesity, chronic motor axonopathy, moderate sensory neuropathy at the upper limbs, mild neurogenic atrophy. | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 30, Age of Patient: 33, Age of Death: n/a |
| Reference: | Janssen et al, 2016; | | Description: | Focal seizures with left visual field symptoms and motor signs. Neurological examination disclosed PEO, dysarthria, decreased reflexes, loss of proprioception distally in the legs, discrete pyramidal signs and appendicular ataxia on the left side with an ataxic gait. Mild cognitive decline was present. infratentorial white matter lesions, axonal polyneuropathy. negative myoclonus. | | Mutations: | W748S, W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: 26, Age of Patient: 28, Age of Death: 33.5 |
Back to top | Reference: | Janssen et al, 2016; | | Description: | Migraine, ataxia, polyneuropathy. focal seizures with visual symptoms and motor signs, and secondary generalized tonic-clonic seizure. Clinical neurological examination revealed an ataxic gait and areflexia with sensory loss. Convulsive status epilepticus. Cerebral MRI revealed a left occipital and left thalamic lesion. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | juvenile | | Age of Onset: 17, Age of Patient: 20, Age of Death: 23 |
| Reference: | Janssen et al, 2016; | | Description: | Presented with status epilepticus, occipital lobe seizures. Further neurologic examination showed ataxic gait and appendicular ataxia. chronic sensory axonal polyneuropathy. Cognitive decline. At follow-up consultation 6 months after the initial presentation, she had not experienced any more seizures. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | juvenile | | Age of Onset: 18, Age of Patient: 18.7, Age of Death: n/a |
| Reference: | Simonati et al, 2003b; | | Description: | epilepsia partialis continua at 10 months, pyramido-cerebellar syndrome 19 months, myoclonic seizures when 4.5 years old. myoclonic jerks. At the age of 8 years, a brain CT scan showed atrophy of the right occipital cortex. signs and symptoms of liver dysfunction. chronic progressive liver disease. diffuse cortical atrophy. myoclonic epilepsy became resistant to any medication, myoclonic status along with episodes of respiratory failure. His clinical condition rapidly worsened and led to a vegetative state. | | Mutations: | L244P, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 0.8, Age of Patient: 9, Age of Death: 15 |
| Reference: | Simonati et al, 2003b; | | Description: | myoclonic fits of the right limbs and then a dystonic-ataxic syndrome at the age of 5 months. He had a rapid clinical deterioration. At age 6 months, a brain CT scan showed diffuse cortical atrophy. acute hepatic failure. | | Mutations: | L244P, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 0.42, Age of Patient: n/a, Age of Death: 1.1 |
| Reference: | Stewart et al, 2009; | | Description: | Ataxia, PEO, axonal neuropathy, cerebellar atrophy. 3% COX deficient fibers. | | Mutations: | G746S, G848S | | SNPs: | E1143G | | Age group: | juvenile | | Age of Onset: 16, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Hisama et al, 2005; | | Description: | Presented with ataxia and transient diplopia at age 25 and showed ptosis, external ophthalmoplegia, nystagmus, had a mild tremor of outstretched hands, marked dysmetria, Romberg sign, severe ataxia, was unable to stand on one foot, multiple mtDNA deletions in muscle. | | Mutations: | A889T | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 25, Age of Death: n/a |
| Reference: | Hisama et al, 2005; | | Description: | presented at age 7 years with gait difficulty and pes cavus. He developed upper extremity tremor, scanning speech, and horizontal nystagmus by age 15. at age 20 showed ptosis, external ophthalmoplegia, mildly decreased vibration and pin prick sensation, intention tremor, absent ankle and brachioradialis reflexes, and ataxic gait. Over the next 5 years, he experienced progression of the ptosis and ophthalmoplegia. The tremor became disabling at rest and with action, and he became wheelchair-dependent. | | Mutations: | A889T | | SNPs: | E1143G | | Age group: | childhood | | Age of Onset: 7, Age of Patient: 20, Age of Death: n/a |
| Reference: | McFarland et al, 2008; | | Description: | Presented with epilepsy following minor head trauma, 2 months after valproate treatment developed persistent vomiting and encephalopathy, deranged liver function tests, prolonged clotting, elevated ammonia, and a high plasma lactate, Hepatic dysfunction progressed, | | Mutations: | A467T, Q879H, T885S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 2, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Hudson et al, 2006; | | Description: | PEO, ataxia, peripheral neuropathy, and episodes of encephalopathy. Asymptomatic mother had also S433C. | | Mutations: | S433C | | SNPs: | E1143G | | Age group: | unknown | | Age of Onset: n/a, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Onset at age 3 with dementia,lactic acidosis, renal tubulopathy, dysmorphic features, cataract, short stature, myopathy. | | Mutations: | G737R, R943C | | SNPs: | E1143G | | Age group: | childhood | | Age of Onset: 3, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Luoma et al, 2007; | | Description: | Parkinsons disease, tremor, rigidity, hypo-/ bradykinesia. Also had Histiocytosis and breast cancer | | Mutations: | | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: 47, Age of Patient: 67, Age of Death: n/a |
|
| W748S | | | | Number of patients: (with W748S) | 181 | | Found together with: | PNF=Putatively Non-Functional enzyme |  | Show Patient Data |
| Patient data are sorted by mutation combination frequency. | Reference: | Blok et al, 2009; | | Description: | Ptosis, CPEO, polyneuropathy, cerebellar ataxia, dysarthria. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 47, Age of Death: n/a |
| Reference: | Kollberg et al, 2006; | | Description: | Started as an ataxic syndrome, Alpers, ragged red fibers, COX-deficient fibers, mtDNA deletions. mtDNA copy numbers 1.6 fold. She was healthy and developed normally until her late preschool years when she developed slowly progressive ataxia. Occasional myoclonic seizures, and at 13 years of age, valproate treatment was started because of occipital seizures. ataxia, dementia, and cortical blindness. She developed refractory seizures, including multifocal EPC, status epilepticus, and myoclonus. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | childhood | | Age of Onset: 5, Age of Patient: 13, Age of Death: 14 |
| Reference: | Kollberg et al, 2006; | | Description: | psychomotor regression, refractory seizures, stroke-like episodes, hepatopathy, and ataxia. COX-deficient muscle fibers, mtDNA deletions, 160% mtDNA copy number. He was normal until 11 years of age when he developed mild tremor and ataxia. At 13 years of age, he reported occasional migraine-like headaches and was found to have mild ataxia and myoclonus. generalized seizures. mild leftsided hemiparesis, external ophthalmoplegia, and peripheral neuropathy. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | childhood | | Age of Onset: 11, Age of Patient: 24, Age of Death: n/a |
| Reference: | Sarzi et al, 2007; | | Description: | Onset with walking difficulties, seizures at 2.5 years, hepatocellular insufficiency after valproate treatment, alpers syndrome, Liver mtDNA depletion. | | Mutations: | W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 2, Age of Patient: n/a, Age of Death: 3 |
Back to top | Reference: | Tzoulis et al, 2006; | | Description: | Presented with epilepsy and headaches, ataxia, status epilepticus, nystagmus, myoclonus, neuropathy. Sodium valproate treatment 6 weeks prior to death. Liver dysfunction, cause of death status epilepticus and liver failure. Hepatic histology showed acute liver necrosis. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | juvenile | | Age of Onset: 19, Age of Patient: n/a, Age of Death: 19 |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with epilepsy and headaches, ataxia, epilepsy, status epilepticus, myoclonus, neuropathy. Sodium valproate treatment 7 weeks prior to death. Liver dysfunction and rising liver enzymes and bilirubin terminally, cause of death status epilepticus. Hepatic histology showed centrolobular degeneration. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | juvenile | | Age of Onset: 14, Age of Patient: n/a, Age of Death: 23 |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with epilepsy, ataxia, nystagmus, neuropathy, ptosis/ PEO onset at age 65 | | Mutations: | W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: 55, Age of Patient: 74, Age of Death: n/a |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with epilepsy, ataxia, status epilepticus, headaches, myoclonus, neuropathy. Sodium valproate treatment for 8 months. Stopped just prior to death. Liver dysfunction, cause of death liver failure. Hepatic histology showed acute liver necrosis. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | childhood | | Age of Onset: 10, Age of Patient: n/a, Age of Death: 10 |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with epilepsy, headaches, epilepsy, status epilepticus, headaches, nystagmus, and neuropathy. Sodium valproate treatment for 2 months then stopped 2 months prior to death. Liver dysfunction, liver transplant 1 month prior to death, cause of death liver failure. Hepatic histology showed acute liver necrosis. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: 20, Age of Patient: n/a, Age of Death: 20 |
Back to top | Reference: | Tzoulis et al, 2006; | | Description: | Presented with epilepsy, status epilepticus, headaches, nystagmus, myoclonus, neuropathy. Sodium valproate treatment 7 weeks prior to death. Liver dysfunction and rising liver enzymes and bilirubin terminally, cause of death status epilepticus. Hepatic histology showed centrolobular degeneration. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | juvenile | | Age of Onset: 15, Age of Patient: n/a, Age of Death: 21 |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with progressive gait unsteadiness, ataxia, headaches, myoclonus, neuropathy, PEO, ptosis onset at age 44 | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: 36, Age of Patient: 50, Age of Death: n/a |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with progressive gait unsteadiness, ataxia, headaches, neuropathy, PEO ptosis onset age 25 | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: 24, Age of Patient: 43, Age of Death: n/a |
| Reference: | Paus et al, 2008; | | Description: | Presented with PEO, mild bilateral ptosis, dysarthria, ataxia, and neuropathy | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: 36, Age of Patient: 37, Age of Death: n/a |
| Reference: | Amiot et al, 2009; | | Description: | Peripheral neuropathy, PEO, ataxia, hearing loss, dysarthria-dysphonia, urinary tract involvement | | Mutations: | A143V, W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: 40, Age of Patient: 49, Age of Death: n/a |
Back to top | Reference: | Palin et al, 2012; | | Description: | No neurological symptoms, but type-2 diabetes and hypertension. | | Mutations: | W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 63, Age of Death: n/a |
| Reference: | Posada et al, 2010; | | Description: | Ataxic sensory neuropathy, dysarthria, progressive ophthalmoplegia, paresthesia of lower extremities, unsteady gait, slurred speech, absent deep tendon reflexes, vibrations, ataxic gait and positive Romberg sign. Multiple mtDNA deletions, a marked decrease in mtDNA copy number. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: 37, Age of Patient: 47, Age of Death: n/a |
| Reference: | Van Goethem et al, 2004; | | Description: | minor tremor of the hands from age 12 years. In his late 20s, he experienced tingling in both hands, followed by tingling in the legs. In the early fourth decade, he became unsteady at walking, especially in the dark. From his early 40s, his speech became slurred (dysarthria). On examination at age 52 years, he had an ataxic gait, and Romberg test was positive. There was dysarthria and bilateral ophthalmoparesis. generalized areflexia. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | childhood | | Age of Onset: 12, Age of Patient: 52, Age of Death: n/a |
| Reference: | Janssen et al, 2016; | | Description: | Migraine, ataxia, polyneuropathy. focal seizures with visual symptoms and motor signs, and secondary generalized tonic-clonic seizure. Clinical neurological examination revealed an ataxic gait and areflexia with sensory loss. Convulsive status epilepticus. Cerebral MRI revealed a left occipital and left thalamic lesion. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | juvenile | | Age of Onset: 17, Age of Patient: 20, Age of Death: 23 |
| Reference: | Janssen et al, 2016; | | Description: | Presented with status epilepticus, occipital lobe seizures. Further neurologic examination showed ataxic gait and appendicular ataxia. chronic sensory axonal polyneuropathy. Cognitive decline. At follow-up consultation 6 months after the initial presentation, she had not experienced any more seizures. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | juvenile | | Age of Onset: 18, Age of Patient: 18.7, Age of Death: n/a |
Back to top | Reference: | Ashley et al, 2008; | | Description: | reported as Alpers, onset at 16 years, presenting encephalopathy with epilepsy, hepatopathy, and movement disorder (ataxia). 108% mtDNA copy number in muscle. | | Mutations: | W748S, W748S | | Age group: | juvenile | | Age of Onset: 16, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Schulte et al, 2009; | | Description: | cerebellar ataxia, dysarthria/dysphagia, sensory neuropathy, PEO, epilepsy, moderate cerebellar atrophy. | | Mutations: | W748S, W748S | | Age group: | adult | | Age of Onset: 22, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Ataxia, peripheral neuropathy, exercise intolerance, easy fatigueability, cerebellar atrophy, abnormal MRI. 82% mtDNA copy number in blood. | | Mutations: | W748S, W748S | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 30, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Headaches/migraines, ataxia, peripheral neuropathy, exercise intolerance, ophthalmoporesis/CPEO, abnormal EMG/NCV, ptosis, constipation, pseudo-obstruction, hearing loss, abnormal BAERS. 83% mtDNA copy number in blood. | | Mutations: | W748S, W748S | | SNPs: | S28C | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 25, Age of Death: n/a |
| Reference: | Winterthun et al, 2005; | | Description: | Ataxia, sensory ataxia, dysarthria, nystagmus, cognitive dysfunction, axonal neuropathy, demyelinating neuropathy. This man presented at age 26 with unsteadiness. Examination showed an ataxic gait, cerebellar dysarthria, myoclonic jerks of his head and facial muscles, mild limitation of horizontal eye movement, and horizontal nystagmus in the direction of gaze, with an additional vertical element when looking down. There were distal amyotrophy, absent reflexes in the legs, and a symmetric loss of all sensory modalities below the knee. Romberg’s test was positive. Episodes of depression. At age 31 he has an almost complete ophthalmoplegia, cerebellar dysarthria, myoclonus involving face and arms, truncal ataxia, and symmetric dysmetria. | | Mutations: | Q497H, Q497H, W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 23, Age of Patient: 38, Age of Death: n/a |
Back to top | Reference: | Winterthun et al, 2005; | | Description: | Headaches, a focal epilepsy with secondary generalisation, occipital epilepsy, dysarthria, nystagmus, cognitive dysfunction. Demyelinating neuropathy, axonal neuropathy. headaches preceded by visual symptoms, nausea, vomiting, and unsteadiness diagnosed as migraine. Shortly after, she had the first of a series of tonic-clonic seizures preceded by headache. Examination showed horizontal and vertical nystagmus, gait ataxia. | | Mutations: | Q497H, Q497H, W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | juvenile | | Age of Onset: 15, Age of Patient: 18, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Onset 33 years with neuropathy, myopathy, SANDO, lactic acidosis, PEO. | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 33, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Milone et al, 2011; | | Description: | Stiffness, cramping of the lower extremities, foot numbness, and poor balance. The past medical history was significant for hypogonadotropic hypogonadism diagnosed 10 years prior. ophthalmoplegia without ptosis, mild lower proximal weakness, Multiple mtDNA deletions detected by PCR in muscle | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 32, Age of Patient: 33, Age of Death: n/a |
| Reference: | Milone et al, 2011; | | Description: | progressive unsteadiness,limb paresthesias, dysarthria, dysphagia, and weakness, ophthalmoparesis,dysarthria, palatal tremor, moderate axial and appendicular ataxia, and distal pan-modality sensory loss | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 32, Age of Patient: 33, Age of Death: n/a |
| Reference: | Pelayo-Negro et al, 2012; | | Description: | ataxia, sensory neuropathy, dysarthria, and external ophthalmoparesis | | Mutations: | W748S, W748S | | Age group: | adult | | Age of Onset: 30, Age of Patient: 51, Age of Death: n/a |
Back to top | Reference: | Tang et al, 2012; | | Description: | migraine, ptosis, PEO, exercise intolerance, sensorimotor peripheral neuropathy, ataxia, pseudoobstruction, constipation, and sensorineural hearing loss, | | Mutations: | W748S, W748S | | SNPs: | S28C | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 25, Age of Death: n/a |
| Reference: | Hakonen et al, 2005; | | Description: | Onset with balance disturbances and neuropathy, Gait and limb ataxia, Dysarthria, restricted eye movements, ptosis, Cognitive impairment, Obesity, cramps, sensory motor polyneuropathy | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 38, Age of Patient: 44, Age of Death: n/a |
| Reference: | Hakonen et al, 2005; | | Description: | Onset with balance disturbances and neuropathy, Gait and limb ataxia, Myoclonus, tremor, Dysarthria, restricted eye movements, Cognitive impairment, Psychiatric symptoms, Obesity, sensory motor polyneuropathy | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 38, Age of Patient: 51, Age of Death: n/a |
| Reference: | Hakonen et al, 2005; | | Description: | Onset with balance disturbances and neuropathy, Gait and limb ataxia, tremor, Involuntary movements, Dysarthria, Dysphagia, Nystagmus, restricted eye movements, Psychiatric symptoms, Muscle strength decreased, Obesity, muscle cramps, amyotrophy, pes cavus, sensory motor polyneuropathy | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 27, Age of Patient: 38, Age of Death: n/a |
| Reference: | Hakonen et al, 2005; | | Description: | Onset with balance disturbances, Gait and limb ataxia, Dysarthria, Dysphagia, Nystagmus, diplopia, Cognitive impairment, Muscle strength decreased, sensory neural hearing deficit, sensory motor polyneuropathy | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 32, Age of Patient: 46, Age of Death: n/a |
Back to top | Reference: | Hakonen et al, 2005; | | Description: | Onset with balance disturbances, Gait and limb ataxia, facial Involuntary movements, Dysarthria, Dysphagia, Nystagmus, other eye-movement abnormalities, Psychiatric symptoms, premature menopause, Obesity, sensory motor polyneuropathy | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 27, Age of Patient: 42, Age of Death: n/a |
| Reference: | Hakonen et al, 2005; | | Description: | Onset with epilepsy, Gait and limb ataxia, Epilepsy, Dysarthria, Nystagmus, diplopia, Psychiatric symptoms, Obesity, sensory motor polyneuropathy | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 24, Age of Patient: 55, Age of Death: n/a |
| Reference: | Hakonen et al, 2005; | | Description: | Onset with epilepsy, Gait and limb ataxia, Epilepsy, Myoclonus, facial Involuntary movements, Dysarthria, Dysphagia, Nystagmus, other eye-movement abnormalities, Cognitive impairment, Psychiatric symptoms, Muscle strength decreased, Obesity | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | juvenile | | Age of Onset: 16, Age of Patient: 37, Age of Death: n/a |
| Reference: | Hakonen et al, 2005; | | Description: | Onset with epilepsy, Gait and limb ataxia, Epilepsy, Myoclonus, head tremor, Dysarthria, other eye-movement abnormalities, Nystagmus, Cognitive impairment, Psychiatric symptoms, cramps, sensory motor polyneuropathy | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 23, Age of Patient: 45, Age of Death: n/a |
| Reference: | Hakonen et al, 2005; | | Description: | Onset with epilepsy, Gait and limb ataxia, Epilepsy, Myoclonus, head tremor, facial Involuntary movements, Dysarthria, Nystagmus, other eye-movement abnormalities, Cognitive impairment, Psychiatric symptoms, cramps | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | juvenile | | Age of Onset: 17, Age of Patient: 45, Age of Death: n/a |
Back to top | Reference: | Hakonen et al, 2005; | | Description: | Onset with epilepsy, Gait and limb ataxia, Myoclonus, Involuntary movements, Dysarthria, Dysphagia, Nystagmus, other eye-movement abnormalities, Cognitive impairment, Obesity, sensory neural hearing deficit, sensory motor polyneuropathy | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | juvenile | | Age of Onset: 19, Age of Patient: 44, Age of Death: n/a |
| Reference: | Hakonen et al, 2005; | | Description: | Onset with headaches, Gait and limb ataxia, Epilepsy, Myoclonus, Involuntary movements, Dysarthria, Dysphagia, Nystagmus, other eye-movement abnormalities, restricted eye movements, ptosis, Cognitive impairment, Psychiatric symptoms, Muscle strength decreased, Obesity | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 28, Age of Patient: 51, Age of Death: n/a |
| Reference: | Hakonen et al, 2005; | | Description: | Onset with neuropathy, Gait and limb ataxia, Dysarthria, Dysphagia, other eye-movement abnormalities, Cognitive impairment, Psychiatric symptoms, Obesity, sensory motor polyneuropathy | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 41, Age of Patient: 58, Age of Death: n/a |
| Reference: | Hakonen et al, 2005; | | Description: | Onset with neuropathy, Gait and limb ataxia, tremor, Dysarthria, Nystagmus, Psychiatric symptoms, Muscle strength decreased, amyotrophy, pes cavus, sensory motor polyneuropathy | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 36, Age of Patient: 51, Age of Death: n/a |
| Reference: | Hakonen et al, 2005; | | Description: | Onset with tremor, Gait and limb ataxia, Epilepsy, Myoclonus, Involuntary movements, Dysarthria, eye muscle weakness, Cognitive impairment, Psychiatric symptoms, Muscle strength decreased. | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | childhood | | Age of Onset: 5, Age of Patient: n/a, Age of Death: 35 |
Back to top | Reference: | Tzoulis et al, 2006; | | Description: | Presented with epilepsy, ataxia, status epilepticus, headaches, myoclonus, neuropathy | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | juvenile | | Age of Onset: 18, Age of Patient: 19, Age of Death: n/a |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with epilepsy, ataxia, status epilepticus, headaches, myoclonus, neuropathy, ptosis/ PEO onset at age 28. Acute liver failure after 4 months of sodium valproate treatment | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | juvenile | | Age of Onset: 19, Age of Patient: 33, Age of Death: n/a |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with epilepsy, ataxia, status epilepticus, headaches, nystagmus, myoclonus, neuropathy, treatment with sodium valproate for 2 months, died 3 years later due to status epilepticus, disseminated intravascular coagulation, liver failure. | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | childhood | | Age of Onset: 10, Age of Patient: n/a, Age of Death: 22 |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with epilepsy, status epilepticus, nystagmus, neuropathy | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | juvenile | | Age of Onset: 17, Age of Patient: 18, Age of Death: n/a |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with migraine like headaches, ataxia, epilepsy, status epilepticus, nystagmus, myoclonus, neuropathy, ptosis/ PEO onset at age 26 | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | juvenile | | Age of Onset: 17, Age of Patient: 38, Age of Death: n/a |
Back to top | Reference: | Tzoulis et al, 2006; | | Description: | Presented with migraine like headaches, ataxia, epilepsy, status epilepticus, nystagmus, myoclonus, neuropathy. Sodium valproate treatment 3 months prior to death. Liver dysfunction and rising liver enzymes and bilirubin terminally, cause of death status epilepticus and multi-organ failure. Acute liver necrosis | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | childhood | | Age of Onset: 8, Age of Patient: n/a, Age of Death: 9 |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with migraine like headaches, ataxia, epilepsy, status epilepticus, nystagmus, neuropathy | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | juvenile | | Age of Onset: 17, Age of Patient: 27, Age of Death: n/a |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with migraine like headaches, ataxia, nystagmus, neuropathy, PEO/ Ptosis onset age >30 | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | childhood | | Age of Onset: 12, Age of Patient: 38, Age of Death: n/a |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with progressive gait unsteadiness and headaches, ataxia, epilepsy, status epilepticus, nystagmus, neuropathy. Died 2 months after treatment with sodium valproate, cause of death status epilepticus | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | juvenile | | Age of Onset: 15, Age of Patient: n/a, Age of Death: 57 |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with progressive gait unsteadiness, ataxia, epilepsy, status epilepticus, headaches, myoclonus, neuropathy, ptosis/ PEO onset age 30. Mild liver dysfunction. Cause of death status epilepticus. | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | childhood | | Age of Onset: 4, Age of Patient: n/a, Age of Death: 30 |
Back to top | Reference: | Tzoulis et al, 2006; | | Description: | Presented with speech delay, ataxia, epilepsy, status epilepticus, and headaches. Liver dysfunction and rising liver enzymes and bilirubin terminally, cause of death status epilepticus, multi-organ failure. Hepatic histology showed marked steatosis. | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | childhood | | Age of Onset: 8, Age of Patient: n/a, Age of Death: 9 |
| Reference: | Johansen et al, 2008; | | Description: | Presented with focal epileptic seizures, external ophthalmoplegia and gait unsteadiness. At age 28, during her first pregnancy, she was admitted with focal epileptic seizures that were highly resistant to treatment. Two months later her symptoms worsened and included myoclonic jerks in the extremities and facial dyskinesias. Treatment with sodium valproate resulted in acute severe hepatic failure and she underwent a successful liver transplantation. At 35 mild cognitive deficit, cerebellar dysarthria, palatal tremor, facial dyskinesias, myoclonus, cerebellar and sensory ataxia and signs of peripheral neuropathy with loss of reflexes and sensory disturbance were also present | | Mutations: | W748S, W748S | | Age group: | juvenile | | Age of Onset: 19, Age of Patient: 35, Age of Death: n/a |
| Reference: | Remes et al, 2008; | | Description: | patient noticed gait and balance difficulties at age 46 years, ataxia, CPEO, cortical and cerebellar atrophy, axonal sensory polyneuropathy, Symmetrical bradykinesia and rigidity, mild tremor, paranoid delusions, During the last 2 years of life he had marked rigidity, dysphagia, myoclonic jerks and dystonia | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 46, Age of Patient: 65, Age of Death: n/a |
| Reference: | Uusimaa et al, 2008; | | Description: | Headache, visual symptoms, migraine-like headache, Athetosis, nystagmus, emiparesis, valproic acid induced Liver failure, sepsis, pancreatitis, status epilepticus | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | juvenile | | Age of Onset: 14, Age of Patient: 15, Age of Death: n/a |
| Reference: | Uusimaa et al, 2008; | | Description: | Migraine like headaches, visual symptoms, focal generalized seizures, severe liver failure, valproate induced liver failure | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | juvenile | | Age of Onset: 15, Age of Patient: n/a, Age of Death: 20 |
Back to top | Reference: | Uusimaa et al, 2008; | | Description: | Seizures, status epilepticus, visual symptoms, nystagmus, valproic acid induced liver failure | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | juvenile | | Age of Onset: 17, Age of Patient: 21, Age of Death: n/a |
| Reference: | Rouzier et al, 2013; | | Description: | Parkinsonism, multiple mtDNA deletions. SANDO, Sensory ataxic neuropathy dysarthria and ophtalmoparesis. | | Mutations: | W748S, W748S | | Age group: | adult | | Age of Onset: 37, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Palin et al, 2012; | | Description: | Gait disturbance since childhood. Early onset suggested anticipation. In his 20's he developed photophobia and general clumsiness and benign paroxysmal positional vertigo. From the age of 37 he has had unspecific sensory polyneuropathy, confirmed by electromyography. He had several simple partial seizures at the age of 39, and has mild anxiety and depression. | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | childhood | | Age of Onset: 5, Age of Patient: 41, Age of Death: n/a |
| Reference: | Palin et al, 2012; | | Description: | developed gait disturbance by the age of 35, progressing to increasing clumsiness in lower extremities, dysarthria, diplopia, and occasional amnesia at the age of 44. She developed ataxia, slight polyneuropathy, and external ophthalmoplegia. At the age of 46 she had slightly increased plasma creatine kinase levels and symmetrical cerebellar peduncular white matter signal intensity increase in brain MRI. The first epileptic seizure, requiring treatment by general anesthesia, oc- curred at the age of 55, after which she was hospitalized permanently. From her 30's, she received psychiatric care due to anxiety and depression. A neuropsychological examination revealed decrease in visual reasoning and memory functions. She deceased at the age of 56 due to pneumonia and pulmonary embolism. | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 35, Age of Patient: n/a, Age of Death: 56 |
| Reference: | Tzoulis et al, 2013; | | Description: | Ataxia, peripheral neuropathy, progressive external ophthalmoplegia (PEO). | | Mutations: | W748S, W748S | | Age group: | childhood | | Age of Onset: 12, Age of Patient: 45, Age of Death: n/a |
Back to top | Reference: | Tzoulis et al, 2013; | | Description: | Epilepsy, stroke-like episodes, ataxia, peripheral neuropathy, progressive external ophthalmoplegia (PEO). | | Mutations: | W748S, W748S | | Age group: | childhood | | Age of Onset: 6, Age of Patient: n/a, Age of Death: 41 |
| Reference: | Tzoulis et al, 2013; | | Description: | Epilepsy, stroke-like episodes, ataxia, peripheral neuropathy, progressive external ophthalmoplegia (PEO). | | Mutations: | W748S, W748S | | Age group: | childhood | | Age of Onset: 12, Age of Patient: n/a, Age of Death: 28 |
| Reference: | Tzoulis et al, 2013; | | Description: | Epilepsy, stroke-like episodes, ataxia, peripheral neuropathy, progressive external ophthalmoplegia (PEO). | | Mutations: | W748S, W748S | | Age group: | juvenile | | Age of Onset: 16, Age of Patient: n/a, Age of Death: 24 |
| Reference: | Tzoulis et al, 2013; | | Description: | Epilepsy, stroke-like episodes, ataxia, peripheral neuropathy, progressive external ophthalmoplegia (PEO). | | Mutations: | W748S, W748S | | Age group: | juvenile | | Age of Onset: 17, Age of Patient: n/a, Age of Death: 43 |
| Reference: | Tzoulis et al, 2013; | | Description: | Epilepsy, stroke-like episodes, ataxia, peripheral neuropathy. | | Mutations: | W748S, W748S | | Age group: | juvenile | | Age of Onset: 17, Age of Patient: 34, Age of Death: n/a |
Back to top | Reference: | Sitarz et al, 2014; | | Description: | Ataxia, neuropathy, PEO, MIRAS. | | Mutations: | W748S, W748S | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 36, Age of Death: n/a |
| Reference: | Tzoulis et al, 2014; | | Description: | Epilepsy, stroke-like episode, Ataxia, Neuropathy | | Mutations: | W748S, W748S | | Age group: | juvenile | | Age of Onset: 15, Age of Patient: n/a, Age of Death: 24 |
| Reference: | Tzoulis et al, 2014; | | Description: | Epilepsy, stroke-like episode, Ataxia, Neuropathy, PEO | | Mutations: | W748S, W748S | | Age group: | childhood | | Age of Onset: 6, Age of Patient: n/a, Age of Death: 41 |
| Reference: | Tzoulis et al, 2014; | | Description: | Epilepsy, stroke-like episode, Ataxia, Neuropathy, PEO. | | Mutations: | W748S, W748S | | Age group: | childhood | | Age of Onset: 12, Age of Patient: n/a, Age of Death: 28 |
| Reference: | Tzoulis et al, 2014; | | Description: | Epilepsy, stroke-like episode, Ataxia, Neuropathy, PEO. | | Mutations: | W748S, W748S | | Age group: | juvenile | | Age of Onset: 15, Age of Patient: n/a, Age of Death: 57 |
Back to top | Reference: | Tzoulis et al, 2014; | | Description: | Epilepsy, stroke-like episode, Ataxia, Neuropathy, PEO. | | Mutations: | W748S, W748S | | Age group: | juvenile | | Age of Onset: 16, Age of Patient: n/a, Age of Death: 24 |
| Reference: | Tzoulis et al, 2014; | | Description: | Epilepsy, stroke-like episode, Ataxia, Neuropathy, PEO. | | Mutations: | W748S, W748S | | Age group: | juvenile | | Age of Onset: 17, Age of Patient: n/a, Age of Death: 43 |
| Reference: | Tzoulis et al, 2014; | | Description: | Epilepsy, stroke-like episode, Ataxia, Neuropathy. | | Mutations: | W748S, W748S | | Age group: | infantile | | Age of Onset: 2, Age of Patient: n/a, Age of Death: 13 |
| Reference: | Schicks et al, 2010; | | Description: | PEO, epilepsy, early-onset cerebellar ataxia. Dysarthria, sensory neuropathy, Cerebellar atrophy. | | Mutations: | W748S, W748S | | Age group: | adult | | Age of Onset: 22, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Schicks et al, 2010; | | Description: | early-onset ataxia, epilepsy, sensory neuropathy. | | Mutations: | W748S, W748S | | Age group: | childhood | | Age of Onset: 12.5, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Synofzik et al, 2010; | | Description: | Dystonia of both arms, with predominant dystonic ulnar deviation of the right upper limb with jerky wrist and finger movements. Also her feet show unpatterend jerky movements, which may be classified as myoclonus but are also similar to limb movements in benign hereditary chorea. external ophthalmoplegia, slowing of voluntary saccades and gait ataxia started. | | Mutations: | W748S, W748S | | Age group: | adult | | Age of Onset: 33, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Synofzik et al, 2010; | | Description: | dystonic ulnar deviation of the left upper limb with distal predominance. She showed intermittent facial and jaw opening dystonia. At rest, she had marked postural instability caused by trunk ataxia, which is aggravated by motor actions like e.g. lifting the upper limbs. severe dysarthria, incomplete horizontal and vertical external ophthalmoplegia and ataxia were observed as clinical features of MIRAS. Patient 2 was only able to stand assisted for a few seconds. | | Mutations: | W748S, W748S | | Age group: | adult | | Age of Onset: 40, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Van Goethem et al, 2004; | | Description: | plurimetabolic syndrome. At age 30 years, he noted disturbed balance, and ataxia and severe axonal neuropathy were diagnosed. On examination at age 32 years, he had slowed ocular pursuit movements but with full range of motion. He had severe gait ataxia, imbalance, and trunk ataxia, as well as clumsiness of the hands. Achilles tendon reflexes were absent. dysarthria, obesity, chronic motor axonopathy, moderate sensory neuropathy at the upper limbs, mild neurogenic atrophy. | | Mutations: | W748S, W748S | | SNPs: | E1143G, E1143G | | Age group: | adult | | Age of Onset: 30, Age of Patient: 33, Age of Death: n/a |
| Reference: | Arkadir et al, 2015; | | Description: | A 30-year-old man with sensorineural hearing loss presented with subacute somnolence, slurred speech, and unsteady gait following treatment with peginterferon a-2b and ribavirin for chronic hepatitis C virus. Examination revealed scanning speech, horizontal nystagmus, gait ataxia, and symmetric hyporeflexia with distal sensory loss. Bilateral hypertrophic olivary degeneration. | | Mutations: | W748S, W748S | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 30, Age of Death: n/a |
| Reference: | Henao et al, 2016; | | Description: | a unique combination of lesions in the thalamus, cerebellum and inferior olivary nucleus. ataxia, dysarthria, external ophthalmoparesis, generalized areflexia, abnormal leg pallesthesia, wide-based gait, positive, Romberg test, dysmetria, and a predominantly left dysdiadochokinesia. distal sensorimotor neuropathy. | | Mutations: | W748S, W748S | | Age group: | adult | | Age of Onset: 24, Age of Patient: 29, Age of Death: n/a |
Back to top | Reference: | Paucar et al, 2016; | | Description: | progressive balance difficulties, impaired gait and coordination (ataxia), slurred speech, diplopia, and hypoacusis, insidious cognitive decline, esophoria. Upon exam, signs of both cerebellar and sensory ataxia (positive Romberg’s sign) as well as chorea, myoclonus, areflexia, and complete loss of vibration sense were found. Bradykinesia and marked postural instability. Eye examination revealed broken up smooth pursuit, nystagmus, mild dysconjugation of lateral eye movements, hypometric saccades and partial restriction of vertical gaze. Psychometric evaluation revealed deficits in information processing speed, working memory, attention, and visuospatial skills. A mild sensorineuronal hearing loss. | | Mutations: | W748S, W748S | | Age group: | adult | | Age of Onset: 48, Age of Patient: 60, Age of Death: n/a |
| Reference: | Janssen et al, 2016; | | Description: | Focal seizures with left visual field symptoms and motor signs. Neurological examination disclosed PEO, dysarthria, decreased reflexes, loss of proprioception distally in the legs, discrete pyramidal signs and appendicular ataxia on the left side with an ataxic gait. Mild cognitive decline was present. infratentorial white matter lesions, axonal polyneuropathy. negative myoclonus. | | Mutations: | W748S, W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: 26, Age of Patient: 28, Age of Death: 33.5 |
| Reference: | Janssen et al, 2016; | | Description: | He presented with two focal seizures with jerking of the right arm and leg. Brain imaging showed a T2-hyperintense lesion in the left frontal lobe. Focal status epilepticus characterized by visual hallucinations in the left visual fields. An appendicular ataxia, nystagmus and areflexia. His level of consciousness deteriorated, evolving into a refractory subtle SE. The patient died eventually after 5 months of continuous epileptic activity due to a septic shock. | | Mutations: | W748S, W748S | | Age group: | juvenile | | Age of Onset: 17, Age of Patient: 20, Age of Death: 20 |
| Reference: | Blok et al, 2009; | | Description: | MELAS-like, features including occipital lobe epilepsy. | | Mutations: | A467T, W748S | | Age group: | juvenile | | Age of Onset: n/a, Age of Patient: 19, Age of Death: n/a |
| Reference: | Blok et al, 2009; | | Description: | Occipital lobe epilepsy, myoclonus, cognitive delay, polyneuropathy, cerebellar ataxia. | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 40, Age of Death: n/a |
Back to top | Reference: | Horvath et al, 2006; | | Description: | Onset at 34 years with PEO, ataxia, dysphagia, myopathy, neuropathy and Diabetes. | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 34, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Schulte et al, 2009; | | Description: | Dysarthria, sensory neuropathy, PEO, minor cognitive dysfunction, depression. | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 34, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Stewart et al, 2009; | | Description: | PEO, ataxia, seizures, ptosis, bilateral deafness, myopathy, dysarthria, peripheral neuropathy, multiple mtDNA deletions. 5% COX deficient fibers, 2% RRF. | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 39, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Autistic features, headaches/migraines, ataxia, seizures, intractable seizure, optic atrophy, abnormal MRI, dystonia, posterior stroke, abnormal EEG. 101% mtDNA copy number in blood. | | Mutations: | A467T, W748S | | Age group: | juvenile | | Age of Onset: n/a, Age of Patient: 18, Age of Death: n/a |
| Reference: | Hansen et al, 2012; | | Description: | gait instability, manual coordination disorder, speech disturbance and secondary generalized motor seizures, tension-type headaches, complete external ophthalmoplegia, cerebellar dysarthria and ataxia, sensory ataxia in Romberg-testing, areflexia of the lower extremities, mild cognitive dysfunction, epileptic seizures, generalised cortical atrophy, sensory axonal neuropathy, dysarthria | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 23, Age of Patient: 48, Age of Death: n/a |
Back to top | Reference: | Kinghorn et al, 2012; | | Description: | Ataxia, dysarthria, suffered two generalized seizures, and developed severe progressive cognitive decline and psychiatric manifestations including visual and auditory hallucinations, bilateral external ophthalmoplegia, bilateral ptosis, and reduced visual acuities, bilateral sensorineural hearing loss, proximal muscle weakness in all four limbs and was areflexic, severe sensory neuropathy and myopathy, | | Mutations: | A467T, W748S | | Age group: | juvenile | | Age of Onset: n/a, Age of Patient: 18, Age of Death: 65 |
| Reference: | Kinghorn et al, 2012; | | Description: | Diplopia, partial ophthalmoparesis in all directions of gaze and dysarthria, sensory neuropathy, COX-deficient fibers, multiple mtDNA deletions, | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 50, Age of Death: n/a |
| Reference: | Pelayo-Negro et al, 2012; | | Description: | ataxia, sensory neuropathy, dysarthria, and external ophthalmoparesis | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 30, Age of Patient: 50, Age of Death: n/a |
| Reference: | Lax et al, 2012a; | | Description: | CPEO, Ptosis, Peripheral neuropathy, COX-deficient fibers, presence of mitochondrial dna deletions in muscle, ataxia, Severe sensory, moderate motor axonal neuronopathy, Distal neurogenic, proximal myopathy | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 41, Age of Patient: 48, Age of Death: n/a |
| Reference: | Lax et al, 2012a; | | Description: | CPEO, Ptosis, Peripheral neuropathy, COX-deficient fibers, ragged red fibers, presence of mitochondrial dna deletions in muscle, Severe sensory and moderate motor neuronopathy, Distal neurogenic change, proximal myopathy | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 34, Age of Patient: 47, Age of Death: n/a |
Back to top | Reference: | Lax et al, 2012a; | | Description: | Peripheral neuropathy, ataxia, epilepsy, COX-deficient fibers, presence of mitochondrial dna deletions in muscle, Moderate sensory neuronopathy | | Mutations: | A467T, W748S | | Age group: | juvenile | | Age of Onset: 16, Age of Patient: 18, Age of Death: n/a |
| Reference: | Lax et al, 2012a; | | Description: | Peripheral neuropathy, ataxia, epilepsy, presence of mitochondrial dna deletions in muscle, | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 20, Age of Patient: 24, Age of Death: n/a |
| Reference: | Paus et al, 2008; | | Description: | episodic headache, cognitive decline, and seizures, repeatedly resulting in generalized status epilepticus, ataxia, sensorimotor peripheral neuropathy, dysarthria, PEO with diplopia, mild bilateral ptosis, epilepsy, cognitive impairment, myoclonus, | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 28, Age of Patient: 39, Age of Death: n/a |
| Reference: | Rouzier et al, 2013; | | Description: | R-EPC (refractory epilepsia partialis continua), axonal neuropathy, cerebellar ataxia, hyperintensity of rolandic, occipital and cerebellar cortex and dentate nucleus. | | Mutations: | A467T, W748S | | Age group: | juvenile | | Age of Onset: 17, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Roshal et al, 2011; | | Description: | Focal parieto-occipital lobe seizures, migraine headaches, cerebellar ataxia, sensory–motor axonal neuropathy, and impairment of visual perception and cognitive function. | | Mutations: | A467T, W748S | | Age group: | juvenile | | Age of Onset: 15, Age of Patient: 16, Age of Death: n/a |
Back to top | Reference: | Tuladhar et al, 2013; | | Description: | abnormal gait, ataxia, PEO, and jerky torticollis, mild cerebellar atrophy, sensory neuronopathy. | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 43, Age of Patient: 45, Age of Death: n/a |
| Reference: | Tzoulis et al, 2013; | | Description: | Ataxia, peripheral neuropathy, progressive external ophthalmoplegia (PEO). | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 24, Age of Patient: 50, Age of Death: n/a |
| Reference: | Tzoulis et al, 2013; | | Description: | Ataxia, peripheral neuropathy, progressive external ophthalmoplegia (PEO). | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 36, Age of Patient: 58, Age of Death: n/a |
| Reference: | Hanisch et al, 2014; | | Description: | Ataxia, ptosis, mild cognitive impairment, sensory neuropathy, motor neuropathy, axonal neuropathy, demyelinating neuropathy. | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 41, Age of Patient: 45, Age of Death: n/a |
| Reference: | Hanisch et al, 2014; | | Description: | Ataxia, ptosis, pareses, diabetes, sensory neuropathy, motor neuropathy, axonal neuropathy, demyelinating neuropathy. | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 49, Age of Patient: 56, Age of Death: n/a |
Back to top | Reference: | Lax et al, 2012b; | | Description: | Ataxia, Epilepsy, Dementia, Encephalopathy, Myoclonus, Fatigue, Depression, Dysarthria, Cardiac failure, cardiomyopathy, Diabetes, arPEO, Ischemic heart disease. | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 20, Age of Patient: 24, Age of Death: n/a |
| Reference: | Sitarz et al, 2014; | | Description: | Ataxia, neuropathy, PEO, MIRAS. | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 39, Age of Death: n/a |
| Reference: | Tzoulis et al, 2014; | | Description: | Epilepsy, stroke-like episode, Ataxia, Neuropathy. | | Mutations: | A467T, W748S | | Age group: | juvenile | | Age of Onset: 13, Age of Patient: n/a, Age of Death: 21 |
| Reference: | Tzoulis et al, 2014; | | Description: | Epilepsy, stroke-like episode, Ataxia, Neuropathy. | | Mutations: | A467T, W748S | | Age group: | juvenile | | Age of Onset: 14, Age of Patient: n/a, Age of Death: 23 |
| Reference: | Janssen et al, 2016; | | Description: | She had occipital lobe and secondarily generalized seizures. In addition, she had PEO, truncal and appendicular ataxia and peripheral neuropathy with diminished vibration sense and areflexia. She reported sporadic occurrence of seizures with a frequency of once in every few months. | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 21, Age of Patient: 47, Age of Death: n/a |
Back to top | Reference: | Ashley et al, 2008; | | Description: | Reported as Alpers, onset at 6 years, presenting encephalopathy with epilepsy, hepatopathy absent, and movement disorder (ataxia). | | Mutations: | G848S, W748S | | Age group: | childhood | | Age of Onset: 6, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Isohanni et al, 2011; | | Description: | Partial status epilepticus, epilepsia partialis, continua, hemiparesis, myoclonus, loss of vision, optical atrophy, pschomotor regression, liver dysfunction, death via pneumonia. | | Mutations: | G848S, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: 11 |
| Reference: | Taanman et al, 2009; | | Description: | Onset at 6.5 years with alpers syndrome and death at 7.8 years. 23% mtDNA copy number in liver. | | Mutations: | G848S, W748S | | SNPs: | E1143G | | Age group: | childhood | | Age of Onset: 6.5, Age of Patient: n/a, Age of Death: 7.8 |
| Reference: | Tang et al, 2011; | | Description: | Headaches/migrains, seizures, intractable seizure, cortical blindness. 32% mtDNA copy number in blood. | | Mutations: | G848S, W748S | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 7, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Hypotonia, myoclonus, or myoclonic seizures, intractable seizure, hepatic failure, abnormal EEG. 38% mtDNA copy number in blood. | | Mutations: | G848S, W748S | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 5, Age of Death: n/a |
Back to top | Reference: | Davidzon et al, 2005; | | Description: | hepatocerebral syndrome, alpers, developmental delay, seizures, and hepatopathy leading to liver failure, liver mtDNA depletion | | Mutations: | G848S, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 0.7, Age of Patient: n/a, Age of Death: 2.5 |
| Reference: | Davidzon et al, 2005; | | Description: | hepatocerebral syndrome, alpers, developmental delay, seizures, and hepatopathy leading to liver failure, liver mtDNA depletion, microvesicular steatosis, | | Mutations: | G848S, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: 6.5 |
| Reference: | Davidzon et al, 2005; | | Description: | hepatocerebral syndrome, alpers, developmental delay, seizures, and hepatopathy leading to liver failure, liver mtDNA depletion, microvesicular steatosis. | | Mutations: | G848S, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 0.1, Age of Patient: n/a, Age of Death: 0.9 |
| Reference: | Davidzon et al, 2005; | | Description: | hepatocerebral syndrome, alpers, developmental delay, seizures, and hepatopathy leading to liver failure, microvesicular steatosis, fibrosis | | Mutations: | G848S, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 0.3, Age of Patient: n/a, Age of Death: 1 |
| Reference: | Nguyen et al, 2005; | | Description: | Refractory seizures, psychomotor regression, liver disease, presenting symptom was status epilepticus, Transient hypoglycemia, Transient lactic acidemia | | Mutations: | G848S, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: 1.7 |
Back to top | Reference: | Wolf et al, 2009; | | Description: | Developemental delay, status epilepticus onset at 6 years 7 months, valproic acid therapy 6 weeks, liver failure with transplant, epilepsia partialis continua, elevated CSF lactate, elevated CSF protein, mtDNA depletion in liver | | Mutations: | G848S, W748S | | Age group: | infantile | | Age of Onset: 6.6, Age of Patient: 7.8, Age of Death: n/a |
| Reference: | Wolf et al, 2009; | | Description: | status epilepticus onset epilepsia partialis continua, elevated CSF lactate, elevated CSF protein, | | Mutations: | G848S, W748S | | Age group: | childhood | | Age of Onset: 10.67, Age of Patient: n/a, Age of Death: 11.5 |
| Reference: | Saneto et al, 2010; | | Description: | seizure onset at 2 years, complex partial seizure and epilepsia partialis continua myoclonus, Truncal ataxia, intention tremor | | Mutations: | G848S, W748S | | Age group: | infantile | | Age of Onset: 2, Age of Patient: 4, Age of Death: n/a |
| Reference: | Hunter et al, 2011; | | Description: | Myoclonic Seizures, Generalized tonic-clonic seizures, Focal motor seizures, Epilepsia partialis continua, Developmental Delay or Regression, ataxia, Abnormal Liver Enzymes, Serum Lactate, Alpers | | Mutations: | G848S, W748S | | Age group: | childhood | | Age of Onset: 6, Age of Patient: n/a, Age of Death: 6.75 |
| Reference: | Hunter et al, 2011; | | Description: | Myoclonic Seizures, Generalized tonic-clonic seizures, Focal motor seizures, Epilepsia partialis continua, Developmental Delay or Regression, ataxia, Abnormal Liver Enzymes, Serum Lactate, liver mtDNA depletion, Alpers | | Mutations: | G848S, W748S | | Age group: | infantile | | Age of Onset: 2, Age of Patient: n/a, Age of Death: 6.33 |
Back to top | Reference: | Wong et al, 2008; | | Description: | Hearing loss, Ataxia Neuropathy Spectrum, Seizures, dementia, developmental delay | | Mutations: | A467T, Q497H, W748S | | SNPs: | E1143G | | Age group: | juvenile | | Age of Onset: 17, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | developmental delay, dementia, seizures, Alpers, Family history of Alpers syndrome. | | Mutations: | G848S, Q497H, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Brunetti-Pierri et al, 2008; | | Description: | He exhibited a very similar phenotype to his sister, with a catastrophic onset of intractable seizures at the age of 9 months, progressive encephalopathy with epilepsia partialis continua and myoclonic jerks, and developmental regression. MRI imaging was not performed. | | Mutations: | G848S, Q497H, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: 1.25, Age of Death: n/a |
| Reference: | Brunetti-Pierri et al, 2008; | | Description: | He exhibited a very similar phenotype to his sister, with a catastrophic onset of intractable seizures at the age of 9 months, progressive encephalopathy with epilepsia partialis continua and myoclonic jerks, and developmental regression. MRI imaging was not performed. | | Mutations: | G848S, Q497H, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: 1.25, Age of Death: n/a |
| Reference: | Brunetti-Pierri et al, 2008; | | Description: | The index patient presented at the age of 8 months with microcephaly, seizures, and developmental delay. From the time of the first observation she rapidly developed epilepsia partialis continua, multifocal myoclonic jerks, and progressive psychomotor retardation. MRI performed at 8 months was generally unremarkable. By 9 months of age the brain MRI showed signifi cant changes including a diffuse severe cerebral and cerebellar atrophy involving gray and white matter. | | Mutations: | G848S, Q497H, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 0.6, Age of Death: 1 |
Back to top | Reference: | Ferrari et al, 2005; | | Description: | An apparently myogenic torticollis was noticed after a few months of life; at 10 months, he had two episodes of sudden head drop, followed by the onset of psychomotor arrest/ regression, hypotonia, ataxia and focal myoclonus of the right upper limb. In the following weeks, the myoclonus became subcontinuous, and was associated with rapidly progressive, severe generalized hypotonia and weakness requiring ventilatory assistance. Seizures were partially controlled with a combination of topiramate, clobazam and phenylbarbiturate. symmetrical lesions of the basal ganglia, thalami, cerebellar dentate nuclei and left occipital cortical and subcortical regions. abnormal accumulation of lactic acid in the putamen and a reduction of the N-acetyl aspartate. cholestatic jaundice, hypoglycaemia and hypocoagulation. The diagnostic conclusion was Alpers syndrome. | | Mutations: | W748S | | SNPs: | E1143G, PNF | | Age group: | infantile | | Age of Onset: 0.2, Age of Patient: n/a, Age of Death: 2.5 |
| Reference: | Ferrari et al, 2005; | | Description: | disease-free interval in the first 4 months after birth, followed by persistent vomiting, epileptic crises with loss of eye contact, and myoclonus in the upper limbs with secondary generalization and loss of consciousness. Persistent myoclonus in the upper limbs was associated with profound hypotonia and loss of eye contact. cortical atrophy and leukoencephalopathy in the subcortical areas of the frontal lobes. progressive hepatic failure with enlarged liver, hypoalbuminaemia and predominantly cholestatic jaundice led to the death of the child at 8 months of age. | | Mutations: | W748S | | SNPs: | E1143G, PNF | | Age group: | infantile | | Age of Onset: 0.3, Age of Patient: n/a, Age of Death: 0.67 |
| Reference: | Sarzi et al, 2007; | | Description: | Myoclonus, epilepsy, Psychomotor retardation, Alpers syndrome, steatosis | | Mutations: | W748S | | SNPs: | E1143G, PNF | | Age group: | infantile | | Age of Onset: 0.25, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Muscle weakness, respiratory failure, Alpers, Lactic acidosis, Seizures, Demential , developmental delay | | Mutations: | W748S | | SNPs: | E1143G, PNF | | Age group: | childhood | | Age of Onset: 4, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Wolf et al, 2009; | | Description: | Alpers. At 7 months she developed focal clonic Status epilepticus. SE subsided slowly after a continuous infusion of valproic acid. she developed severe muscular hypotonia and feeding difficulties necessitating nasogastric feeding. Twelve weeks after disease onset, she was readmitted with vomiting and pneumonia. Infantile scoliosis. | | Mutations: | W748S | | SNPs: | PNF | | Age group: | infantile | | Age of Onset: 0.58, Age of Patient: n/a, Age of Death: 0.83 |
Back to top | Reference: | Mousson de Camaret et al, 2011 | | Description: | At 16 months of age, he started a severe status epilepticus. Cerebral MRI showed brain cortical atrophy. he developed intractable epileptic encephalopathy associated with generalized hypotonia and cortical blindness. diffuse hypotonia, lethargy and vomiting. During the two following years, epilepsy was always very active including spasms and myoclonic jerks. progressive hepatic failure. Alpers. | | Mutations: | W748S | | SNPs: | E1143G, PNF | | Age group: | infantile | | Age of Onset: 1.33, Age of Patient: n/a, Age of Death: 4.17 |
| Reference: | Rouzier et al, 2013; | | Description: | R-EPC (refractory epilepsia partialis continua), delayed psychomotor development. Has unrelated healthy parents. Pregnancy and birth were unremarkable. She had had a moderately delayed psychomotor development with sitting posture at 9 months of age and walking at 20 months of age. At 3 years old, she presented with acute partial status epilepticus. After this first episode, she developed refractory EPC with neurological regression leading to a severe encephalopathy. At 4 years, she was unable to hold her head up or to sit and had poor contact. Muscle biopsy showed lipid accumulation and spectrophotometric measurements of the indivi- dual RC complexes revealed CIII deficiency, associated with decreased activitiy of complexes II and V. In the liver, specific activity of complexes I, III and V was also affected. mtDNA depletion was identified by qPCR (25% of the mean normal control amount of mtDNA relative to nuclear DNA in the muscle and 30% in the liver). Sequencing identified only one heterozygous mutation (p.Trp748Ser) in POLG, inherited from the mother. | | Mutations: | W748S | | SNPs: | PNF | | Age group: | infantile | | Age of Onset: 2, Age of Patient: 4, Age of Death: n/a |
| Reference: | Kaliszewska et al, 2015; | | Description: | a psychomotor retardation and muscle hypotonia were observed since 2 months of age, and autistic behaviour was observed during early childhood. At the age of 3 years, the patient was given valproic acid due to episodes of unconsciousness and abnormal EEG pattern and responded suddenly with an acute liver failure. | | Mutations: | PNF, W748S | | Age group: | infantile | | Age of Onset: 1.67, Age of Patient: n/a, Age of Death: 3.5 |
| Reference: | Ashley et al, 2008; | | Description: | mtDNA depletion in muscle, Epilepsy, hepatopathy, movement disorder | | Mutations: | T914P, W748S | | Age group: | infantile | | Age of Onset: 1.1, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Ashley et al, 2008; | | Description: | reported as Alpers, onset at 4 years, presenting encephalopathy with epilepsy, hepatopathy, and movement disorder (ataxia). 15% mtDNA copy number in liver. | | Mutations: | T914P, W748S | | Age group: | childhood | | Age of Onset: 4, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Isohanni et al, 2011; | | Description: | developmental delay, vomiting, partial status epilepticus, epilepsia partialis continua, myoclonus, ataxia, alive at age 3. | | Mutations: | T914P, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 2.5, Age of Patient: 3, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Age 0.8, developmental delay, hypotonia, myoclinic seizures, spasticity, dystonia, exercise intolerance, GI reflux, abnormal EEG, elevated transaminases, increased signal basal ganglia, abnormal MRI, lactic acidosis, high CSF lactate. | | Mutations: | T914P, W748S | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 0.8, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Seizures. 98% mtDNA copy number in blood. | | Mutations: | T914P, W748S | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 5, Age of Death: n/a |
| Reference: | Hinnell et al, 2012; | | Description: | Presented with generalized epilepsy and mild long-standing learning difficulties requiring special education, developed liver failure after taking valproate, migraines, right-sided epilepsia partialis continua, myoclonic arm jerks, a pancerebellar syndrome, and progressive cognitive impairment. mild external ophthalmoplegia, saccadic pursuit, writhing tongue movements, dysarthria, bilateral dysdiadochokinesis and dysmetria, mild sensory axonal peripheral neuropathy, ptosis | | Mutations: | T914P, W748S | | Age group: | juvenile | | Age of Onset: 15, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Rouzier et al, 2013; | | Description: | Refractory generalized epilepsy with status epilepticus, hepatic cholestasis and cytolysis, proximal tubulopathy, hyperintensity of thalamus. mtDNA depletion. | | Mutations: | T914P, W748S | | Age group: | childhood | | Age of Onset: 6, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Ashley et al, 2008; | | Description: | reported as Alpers, onset at 1 years, presenting encephalopathy with epilepsy, hepatopathy, and movement disorder (ataxia). 32% mtDNA copy number in liver. | | Mutations: | R852C, W748S | | SNPs: | G11D | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Stewart et al, 2009; | | Description: | Alpers, Seizures, Epilepsia partialis continua, liver failure, stroke-like episode. | | Mutations: | R852C, W748S | | SNPs: | E1143G, G11D | | Age group: | infantile | | Age of Onset: 1.33, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Naess et al, 2009; | | Description: | Developmental retardation, Hypotonia, ataxia, seizures myoclonus, abnormal eye movement, gastrointestinal problems, hepatic involvement | | Mutations: | R852C, W748S | | SNPs: | G11D | | Age group: | infantile | | Age of Onset: 0.3, Age of Patient: n/a, Age of Death: 1.1 |
| Reference: | Vasta et al, 2012; | | Description: | PEO, alpers disease, ataxia, seizure, hypotonia, developemental delay, | | Mutations: | R852C, W748S | | SNPs: | E1143G, G11D | | Age group: | infantile | | Age of Onset: 0.001, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Sofou et al, 2013; | | Description: | Alpers/ Alpers–Huttenlocher. | | Mutations: | Q497H, R852C, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Tzoulis et al, 2009; | | Description: | PEO, ptosis, muscle fatigue, diplopia, a mild external ophthalmoplegia affecting horizontal, but not vertical gaze. Tendonreflexes were diminished in the lower extremities and a distal sensory defect in stocking distribution was seen. | | Mutations: | P587L, W748S | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 45, Age of Patient: 50, Age of Death: n/a |
| Reference: | Ylönen et al, 2013; | | Description: | Akinetic-rigid Parkinsons. impaired balance, freezing, and increased salivation. Constipation and muscle cramps. He had arterial hypertension and mild normocytic anemia. cytochrome c oxidase-negative muscle fibers. | | Mutations: | P587L, W748S | | SNPs: | E1142G, T251I | | Age group: | adult | | Age of Onset: 49, Age of Patient: 65, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Peripheral neuropathy, CPEO, ptosis, COX deficiency, ragged red fibers, headaches/migraines, diarrhoea, lactic acidosis, hypotonia, ataxia, myoclonic seizures, exercise intolerance, muscle weakness, muscle cramps after exercise, optic atrophy, easy fatigability. 61% mtDNA copy number in muscle, 61% mtDNA copy number in blood. | | Mutations: | R953C, W748S | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 51, Age of Death: n/a |
| Reference: | Tang et al, 2012; | | Description: | Exercise intolerance, muscle weakness, persistent diarrhea, intermittent vomiting, bilateral ptosis, cognitive impairment with poor recall and expressive language difficulty, proximal myopathy, axonal sensorimotor peripheral neuropathy | | Mutations: | R953C, W748S | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 50, Age of Death: n/a |
| Reference: | Nolte et al, 2013; | | Description: | Generalized tonic-clonic seizures which, within 3 days, evolved to epilepsia partialis continua (EPC) with continuous left-sided myoclonic jerks. Multifocal brain lesions and global brain atrophy. | | Mutations: | R627W, W748S | | Age group: | juvenile | | Age of Onset: 13, Age of Patient: 16, Age of Death: 17 |
Back to top | Reference: | Nolte et al, 2013; | | Description: | This previously healthy 17-year-old patient developed repeated complex partial seizures starting with visual sensations, as well as myoclonic jerks of her right arm that rapidly evolved to generalized tonic-clonic seizures. Repeated complex partial seizures and considerable mental impairment. | | Mutations: | R627W, W748S | | Age group: | juvenile | | Age of Onset: 17, Age of Patient: 17, Age of Death: n/a |
| Reference: | Simonati et al, 2003b; | | Description: | epilepsia partialis continua at 10 months, pyramido-cerebellar syndrome 19 months, myoclonic seizures when 4.5 years old. myoclonic jerks. At the age of 8 years, a brain CT scan showed atrophy of the right occipital cortex. signs and symptoms of liver dysfunction. chronic progressive liver disease. diffuse cortical atrophy. myoclonic epilepsy became resistant to any medication, myoclonic status along with episodes of respiratory failure. His clinical condition rapidly worsened and led to a vegetative state. | | Mutations: | L244P, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 0.8, Age of Patient: 9, Age of Death: 15 |
| Reference: | Simonati et al, 2003b; | | Description: | myoclonic fits of the right limbs and then a dystonic-ataxic syndrome at the age of 5 months. He had a rapid clinical deterioration. At age 6 months, a brain CT scan showed diffuse cortical atrophy. acute hepatic failure. | | Mutations: | L244P, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 0.42, Age of Patient: n/a, Age of Death: 1.1 |
| Reference: | Naimi et al, 2006; | | Description: | SCAE, age of onset 20, death at 27, ptosis and multiple deletions in muscle. | | Mutations: | L304R, W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: 20, Age of Patient: n/a, Age of Death: 27 |
| Reference: | Ashley et al, 2008; | | Description: | Encephalopathy onset 16 years of age, presented with epilepsy. | | Mutations: | P587L, P589L, W748S | | Age group: | juvenile | | Age of Onset: 16, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Isohanni et al, 2011; | | Description: | epilepsy, vomiting, lowered consciousness, partial status epilepticus, epilepsia partialis continua, myoclonus, pschomotor regression, liver failure before valproate treatment, death via pneumonia. 72-76% mtDNA copy number in muscle. | | Mutations: | R807C, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: 3 |
| Reference: | Spinazzola et al, 2009; | | Description: | Onset of Alpers at 3 months, death at 19 months. Bilateral lesions of thalami. | | Mutations: | D930N, W748S | | Age group: | infantile | | Age of Onset: 0.3, Age of Patient: n/a, Age of Death: 1.5 |
| Reference: | Tang et al, 2011; | | Description: | Alpers. 46% mtDNA copy number in blood. | | Mutations: | R852H, W748S | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 3, Age of Death: n/a |
| Reference: | Taanman et al, 2009; | | Description: | Hypotonia at 8 weeks, seizures, hepatopathy, lactic acidemia, died of liver failure. 9% mtDNA copy number in muscle, 11% mtDNA copy number in liver. | | Mutations: | H1110Y, W748S | | SNPs: | E1143G, Q1236H | | Age group: | infantile | | Age of Onset: 0.1, Age of Patient: n/a, Age of Death: 0.8 |
| Reference: | Tang et al, 2011; | | Description: | Seizures, ptosis, hepatic failure, ketosis, lactic acidosis, organic aciduria/tiglyglycine, elevated pyruvate, microcephaly, abnormal MRI. 33% mtDNA copy number in muscle, 46% mtDNA copy number in blood, ETC low. | | Mutations: | G888D, W748S | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 2, Age of Death: n/a |
Back to top | Reference: | Tzoulis et al, 2009; | | Description: | PEO. Primary hypothyroidism and bilateral hearing loss of uncertain duration. She had been operated for bilateral ptosis at 75 years of age and presented to us with 2–3 years of worsening diplopia, gait unsteadiness and paresthaesiae in the distal lower limbs. asymmetrical ptosis and nearly complete external ophthalmoplegia with loss of convergence, oculocephalic reflex and Bell\'s reflex. She had symmetrical distal sensory loss in the lower limbs and absence of Achilles reflexes. | | Mutations: | G737R, W748S | | Age group: | adult | | Age of Onset: 75, Age of Patient: 86, Age of Death: n/a |
| Reference: | Zsurka et al, 2008; | | Description: | Epilepsy, seizures, VPA induced liver failure. | | Mutations: | L752P, W748S | | SNPs: | E1143G | | Age group: | childhood | | Age of Onset: 7, Age of Patient: n/a, Age of Death: 10 |
| Reference: | Kurt et al, 2010; | | Description: | Psychomotor delay, status epilepticus, liver disease, lactic acidosis, ADHD, mental retardation. | | Mutations: | P1073L, W748S | | Age group: | infantile | | Age of Onset: 0.01, Age of Patient: n/a, Age of Death: 13 |
| Reference: | Kollberg et al, 2006; | | Description: | Alpers, age of onset 6 months, age of death 13 months. 41% mtDNA copy number. During the first 6 months of life, he was considered healthy, but was irritable with colic-like symptoms. failure to thrive, delayed motor development. myoclonus. leftsided hemiparesis. | | Mutations: | R232H, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: n/a, Age of Death: 1.08 |
| Reference: | Komulainen et al, 2010; | | Description: | mental retardation, psychiatric symptoms, mild bilateral ptosis and epilepsy, Seizures occurred at age 11 years, and focal generalized epilepsy was diagnosed. | | Mutations: | W748S | | SNPs: | E1143G, R722H | | Age group: | childhood | | Age of Onset: 11, Age of Patient: 22, Age of Death: n/a |
Back to top | Reference: | Lax et al, 2012a; | | Description: | CPEO, Ptosis, Peripheral neuropathy, COX-deficient fibers, ragged red fibers, presence of mitochondrial dna deletions in muscle, epilepsy, Severe sensory and moderate motor neuronopathy, Distal neurogenic change, proximal myopathy | | Mutations: | R1096C, W748S | | Age group: | adult | | Age of Onset: 25, Age of Patient: 49, Age of Death: n/a |
| Reference: | Kollberg et al, 2006; | | Description: | Ragged Red Fibers, COX-Deficient Fibers. mtDNA depletion. He was healthy and his development was normal until 5 months of age. He developed failure to thrive and muscular hypotonia. At 4 years of age, he developed myoclonus, which was first limited to the right eye and the right side of the mouth, but then progressed to epilepsia partialis continua of the entire left side of the body without loss of consciousness. He has developed a complex movement disorder and cognitive impairment, but contact, speech, and memory functions have been retained. He also had ptosis and uncontrolled and uncoordinated movements, especially in his arms. | | Mutations: | E1163G, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 0.417, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Ylönen et al, 2013; | | Description: | late-onset parkinsons. Tremor. Cardiac arrhythmia. | | Mutations: | W748S | | SNPs: | E1143G, R993C | | Age group: | adult | | Age of Onset: 72, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Kaliszewska et al, 2015; | | Description: | Alpers syndrome. At the age of 12 months, he became progressively ataxic and recurrent convulsions appeared, developing quickly to severe epileptic encephalopathy. Fulminant liver failure. Severe mitochondrial depletion found in the liver tissue on autopsy. | | Mutations: | R869X, W748S | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: 2.5 |
| Reference: | Kaliszewska et al, 2015; | | Description: | SANDO. In spite of normal developmental milestones, the patient started to have walking difficulties at the age of 13 years. Her condition deteriorated, and she developed ataxic gait and dysarthria. Two years later, she developed action-exacerbated myoclonus. Nerve conduction studies showed sensorymotor polyneuropathy of lower limb nerves. | | Mutations: | V1106A, W748S | | Age group: | juvenile | | Age of Onset: 13, Age of Patient: 15, Age of Death: n/a |
Back to top | Reference: | Nicastro et al, 2015; | | Description: | A 52-year-old male patient, treated for hypertension and diabetes. Progressive ataxia with palatal tremor (PAPT). progressive gait and balance difficulties since two years. audible ear click and oscillopsia. On neurological examination, he had pendular vertical nystagmus, dysarthria, kinetic and static ataxia with severe postural instability, as well as palatal tremor. He became wheelchair-bound within 4 years after the first symptoms. Family history was unremarkable on the maternal side and unknown on the paternal side. The mother of the patient died at the age of 86 without evidence of neurologic disease. | | Mutations: | I1185N, W748S | | Age group: | adult | | Age of Onset: 50, Age of Patient: 52, Age of Death: n/a |
| Reference: | Sarzi et al, 2007; | | Description: | Feeding difficulties 2 days after birth and trunk hypotonia, dysmorphy, hypotonia, coloboma, heart failure at 2 months, liver insufficiency at 4 months. Liver and muscle mtDNA depletion. | | Mutations: | A467T, K561M, W748S | | Age group: | infantile | | Age of Onset: 0.001, Age of Patient: n/a, Age of Death: 0.5 |
|
|
|
|
|
The following information is based on existing patient data and pathogenic cluster assignment.
Pathogenicity information for a patient with mutation W748S and a cluster 1 mutation: Age of onset information is extracted from a total of 37 patients and/ or patient families. | Age of onset | | |
37- 19- | 21
| 10
| 2
| 4
| | | infantile | childhd | juvenile | adult | | | 57% | 27% | 5% | 11% | |
All mutations mapping within the pathogenic clusters are at high risk for pathogenicity. In general, a patient must have a pathogenic mutation in both of his/ her POLG genes to develop a POLG-related syndrome.  | Symptoms described in patients with W748S-cluster1 mutations | | Symptoms in patients with combination
W748S:cluster1 | | Developmental delay | 45.9% | | Epilepsy | 40.5% | | Movement disorder (ataxia) | 40.5% | | Encephalopathy | 40.5% | | Liver failure | 29.7% | | Myoclonic seizures | 27.0% | | Epilepsia partialis | 27.0% | | Hypotonic | 18.9% | | Status epilepticus | 16.2% | | Lactic acidosis | 13.5% | | Ptosis | 13.5% | | Peripheral neuropathy | 10.8% | | Hepatocerebral | 10.8% | | Dysarthria | 10.8% | | Intractable seizure | 8.1% | | Exercise intolerance | 8.1% | | PEO | 8.1% | | Headache/ migraine | 8.1% | | Ragged red fibers | 5.4% | | Muscle weakness | 5.4% | | Liver dysfunction | 5.4% | | Vomiting | 5.4% | | Tremor | 5.4% | | +24 other symptoms in under 5.0% of the patients |
| | Data gathered from clinical descriptions for 37 patients |
| Symptoms by group | | Seizures | 81.1% | | Developmental Delay | 54.1% | | Hepatopathy | 43.2% | | Alpers syndrome | 40.5% | | Ataxia | 40.5% | | CNS symptoms | 18.9% | | CPEO | 18.9% | | Hypotonia | 18.9% | | Neuropathy | 13.5% | | Other | 13.5% | | GI symptoms | 10.8% | | Myopathy | 10.8% | | Migraines | 8.1% | | Unknown | 2.7% |
| | [Show grouping information] |
|  | Symptoms described in patients with cluster1-cluster5 mutations | |
| Symptoms in patients with combination
cluster1:cluster5 | | Movement disorder (ataxia) | 41.2% | | Developmental delay | 37.3% | | Encephalopathy | 33.3% | | Epilepsy | 31.4% | | Liver failure | 23.5% | | Myoclonic seizures | 19.6% | | Epilepsia partialis | 19.6% | | Ptosis | 15.7% | | PEO | 15.7% | | Lactic acidosis | 13.7% | | Status epilepticus | 13.7% | | Hypotonic | 13.7% | | Headache/ migraine | 11.8% | | Dysarthria | 11.8% | | Peripheral neuropathy | 9.8% | | Intractable seizure | 7.8% | | Stroke | 7.8% | | Hepatocerebral | 7.8% | | Muscle weakness | 5.9% | | Exercise intolerance | 5.9% | | Vomiting | 5.9% | | +37 other symptoms in under 5.0% of the patients |
| | Data gathered from clinical descriptions for 51 patients |
| Symptoms by group | | Seizures | 64.7% | | Developmental Delay | 47.1% | | Ataxia | 43.1% | | Hepatopathy | 37.3% | | Alpers syndrome | 31.4% | | CNS symptoms | 27.5% | | CPEO | 25.5% | | Neuropathy | 17.6% | | Other | 15.7% | | Hypotonia | 13.7% | | Migraines | 11.8% | | Myopathy | 11.8% | | GI symptoms | 9.8% | | Unknown | 3.9% |
| | [Show grouping information] |
|
|
|