Mutation Query
| | | | Allele 1: | G737R | | Allele 2: | R853W | Allelic information known | | Refine query |
|
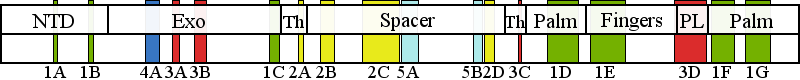
| | | Residue G737 | | Cluster assignment: | | | Cluster description: | Putative protein-protein interactions | | Subcluster: | 5B (residues 737-749) | | Subcluster description: | Located in the periphery of the IP subdomain of the spacer domain, distant from the DNA binding channel | | POLG domain: | Spacer domain |
| Residue R853 | | Cluster assignment: | | | Cluster description: | Polymerase active site and environs | | Subcluster: | 1D (residues 848-895) | | Subcluster description: | This subcluster forms a large portion of the pol active site and contains two highly conserved motifs that are found in all family A polymerases: the RR loop (motif 2) and motif A (Loh and Loeb, 2005). | | POLG domain: | Polymerase domain |
|
|
Mutation Information
|
| G737R | | | | Number of patients: (with G737R) | 13 | | Found together with: | |  | Show Patient Data |
| Patient data are sorted by mutation combination frequency. | Reference: | Wong et al, 2008; | | Description: | Onset 60 years with ataxia, neuropathy, myopathy, hearing loss, cerebellar atrophy, SANDO/SCAE, PEO. | | Mutations: | A467T, G737R | | Age group: | adult | | Age of Onset: 60, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Milone et al, 2011; | | Description: | progressive sensory ataxic neuropathy, ophthalmoparesis, cerebellar ataxia, limb weakness, muscle cramps, sensory hearing loss, dysarthria, dysphagia, constipation, and memory loss, brain MRI was abnormal with evidence of generalized cortical and cerebellar atrophy, evidence of a length-dependent sensory > motor polyneuropathy of axonal type, Multiple mtDNA deletions detected by PCR in blood | | Mutations: | A467T, G737R | | Age group: | adult | | Age of Onset: 53, Age of Patient: 58, Age of Death: n/a |
| Reference: | Horvath et al, 2006; | | Description: | Onset at 0.8 years with encephalopathy, liver dysfunction, diagnosed as Alpers. Death at 1 year. | | Mutations: | A767D, G737R | | Age group: | infantile | | Age of Onset: 0.8, Age of Patient: n/a, Age of Death: 1 |
| Reference: | Sitarz et al, 2014; | | Description: | Epilepsy, Myopathy. | | Mutations: | A767D, G737R | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 1, Age of Death: n/a |
Back to top | Reference: | Davidzon et al, 2006; | | Description: | Migraines in childhood, In 20s presented with peripheral sensory neuropathy and parkinsonisms. | | Mutations: | G737R, R853W | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Rempe et al, 2016; | | Description: | parkinsonism with dystonic toe and plantar flexion. Anxiety and generalized muscle weakness. Bilateral hypaesthesia of the lateral bottom of the foot and dorsal forefoot as well as bilateral distal pallhypaesthesia of the legs. Slight bilateral ptosis, slight axonal sensory polyneuropathy. | | Mutations: | G737R, R853W | | Age group: | adult | | Age of Onset: 27, Age of Patient: 32, Age of Death: n/a |
| Reference: | Harrower et al, 2008; | | Description: | Peripheral neuropathy, tremor, ataxia. | | Mutations: | G737R, R232H | | SNPs: | S64L | | Age group: | childhood | | Age of Onset: 10, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Muscle weakness, ptosis, ophthalmoparesis/CPEO. 61% mtDNA copy number in blood. | | Mutations: | G737R, L304R | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 54, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Seizures, intractable seizures, abnormal EEG, abnormal MRI. 135% mtDNA copy number in blood. | | Mutations: | G426S, G737R | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 11, Age of Death: n/a |
Back to top | Reference: | Tang et al, 2011; | | Description: | Deceased at 9 months. 30% mtDNA copy number in muscle, 30% mtDNA copy number in blood, 10% mtDNA copy number in liver. | | Mutations: | A957V, G737R | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 0.4, Age of Death: 0.8 |
| Reference: | Tang et al, 2011; | | Description: | Hypoglycaemia, liver failure, seizures, developmental delay. 65% mtDNA copy number in muscle, ETC low. | | Mutations: | G737R, V855L | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 0.8, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Onset at age 3 with dementia,lactic acidosis, renal tubulopathy, dysmorphic features, cataract, short stature, myopathy. | | Mutations: | G737R, R943C | | SNPs: | E1143G | | Age group: | childhood | | Age of Onset: 3, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Tzoulis et al, 2009; | | Description: | PEO. Primary hypothyroidism and bilateral hearing loss of uncertain duration. She had been operated for bilateral ptosis at 75 years of age and presented to us with 2–3 years of worsening diplopia, gait unsteadiness and paresthaesiae in the distal lower limbs. asymmetrical ptosis and nearly complete external ophthalmoplegia with loss of convergence, oculocephalic reflex and Bell\'s reflex. She had symmetrical distal sensory loss in the lower limbs and absence of Achilles reflexes. | | Mutations: | G737R, W748S | | Age group: | adult | | Age of Onset: 75, Age of Patient: 86, Age of Death: n/a |
|
| R853W | | | | Number of patients: (with R853W) | 2 | | Non-allelic with: | G737R (100%) |  | Show Patient Data |
| Patient data are sorted by mutation combination frequency. | Reference: | Davidzon et al, 2006; | | Description: | Migraines in childhood, In 20s presented with peripheral sensory neuropathy and parkinsonisms. | | Mutations: | G737R, R853W | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Rempe et al, 2016; | | Description: | parkinsonism with dystonic toe and plantar flexion. Anxiety and generalized muscle weakness. Bilateral hypaesthesia of the lateral bottom of the foot and dorsal forefoot as well as bilateral distal pallhypaesthesia of the legs. Slight bilateral ptosis, slight axonal sensory polyneuropathy. | | Mutations: | G737R, R853W | | Age group: | adult | | Age of Onset: 27, Age of Patient: 32, Age of Death: n/a |
|
|
|
|
|
The following information is based on existing patient data and pathogenic cluster assignment.
Pathogenicity information for a patient with mutation G737R and a cluster 1 mutation: Age of onset information is extracted from a total of 6 patients and/ or patient families. | Age of onset | | |
6- 3- | 2
| 3
| 0
| 1
| | | infantile | childhd | juvenile | adult | | | 33% | 50% | 0% | 17% | |
All mutations mapping within the pathogenic clusters are at high risk for pathogenicity. In general, a patient must have a pathogenic mutation in both of his/ her POLG genes to develop a POLG-related syndrome.  | Symptoms described in patients with G737R-cluster1 mutations | | Symptoms in patients with combination
G737R:cluster1 | | Parkinson's disease | 33.3% | | No known symptoms | 16.7% | | Lactic acidosis | 16.7% | | Intractable seizure | 16.7% | | Polyneuropathy | 16.7% | | Sensomotor neuropathy | 16.7% | | Muscle weakness | 16.7% | | Myopathy | 16.7% | | Ptosis | 16.7% | | Liver failure | 16.7% | | Headache/ migraine | 16.7% | | Developmental delay | 16.7% | | Dementia | 16.7% | | Renal tubulopathy | 16.7% |
| | Data gathered from clinical descriptions for 6 patients |
| Symptoms by group | | CNS symptoms | 33.3% | | Developmental Delay | 33.3% | | Hepatopathy | 33.3% | | Myopathy | 33.3% | | Neuropathy | 33.3% | | CPEO | 16.7% | | Migraines | 16.7% | | Other | 16.7% | | Seizures | 16.7% | | Unknown | 16.7% |
| | [Show grouping information] |
|  | Symptoms described in patients with cluster5-cluster1 mutations | |
| Symptoms in patients with combination
cluster1:cluster5 | | Movement disorder (ataxia) | 41.2% | | Developmental delay | 37.3% | | Encephalopathy | 33.3% | | Epilepsy | 31.4% | | Liver failure | 23.5% | | Myoclonic seizures | 19.6% | | Epilepsia partialis | 19.6% | | Ptosis | 15.7% | | PEO | 15.7% | | Lactic acidosis | 13.7% | | Status epilepticus | 13.7% | | Hypotonic | 13.7% | | Headache/ migraine | 11.8% | | Dysarthria | 11.8% | | Peripheral neuropathy | 9.8% | | Intractable seizure | 7.8% | | Stroke | 7.8% | | Hepatocerebral | 7.8% | | Muscle weakness | 5.9% | | Exercise intolerance | 5.9% | | Vomiting | 5.9% | | +37 other symptoms in under 5.0% of the patients |
| | Data gathered from clinical descriptions for 51 patients |
| Symptoms by group | | Seizures | 64.7% | | Developmental Delay | 47.1% | | Ataxia | 43.1% | | Hepatopathy | 37.3% | | Alpers syndrome | 31.4% | | CNS symptoms | 27.5% | | CPEO | 25.5% | | Neuropathy | 17.6% | | Other | 15.7% | | Hypotonia | 13.7% | | Migraines | 11.8% | | Myopathy | 11.8% | | GI symptoms | 9.8% | | Unknown | 3.9% |
| | [Show grouping information] |
|
|
|