Mutation Query
| | | | Allele 1: | L304R | | Allele 2: | G888D | Allelic information known | | Refine query |
|
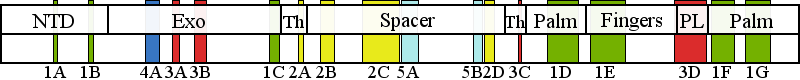
| | | Residue L304 | | Cluster assignment: | | | Cluster description: | Partitioning loop | | Subcluster: | 3B (residues 303-319) | | Subcluster description: | A helix-coil-helix module (residues 295-312) located in the Exo domain that has been termed the "orienter" module | | POLG domain: | Exonuclease domain |
| Residue G888 | | Cluster assignment: | | | Cluster description: | Polymerase active site and environs | | Subcluster: | 1D (residues 848-895) | | Subcluster description: | This subcluster forms a large portion of the pol active site and contains two highly conserved motifs that are found in all family A polymerases: the RR loop (motif 2) and motif A (Loh and Loeb, 2005). | | POLG domain: | Polymerase domain |
|
|
Mutation Information
|
| L304R | | | | Number of patients: (with L304R) | 23 | | Found as the only mutation: | 13% of entries (3 patients) | | Found together with: | |  | Show Patient Data |
| Patient data are sorted by mutation combination frequency. | Reference: | Ashley et al, 2008; | | Description: | Relatively Mild phenotype | | Mutations: | L304R, L304R | | Age group: | childhood | | Age of Onset: 10, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | CPEO, muscle weakness, ptosis, ataxia. 53% mtDNA copy number in blood. | | Mutations: | L304R, L304R | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 10, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | CPEO, ptosis, ataxia, abnormal EMG/NCV, cerebellar atrophy, large mitochondrial proliferation, ragged red fibers, easy fatigability, abnormal muscle histology, hypotonia, lactic acidosis, high CSF lactate, organic aciduria/tiglyglycine, low plasma carnitine. 36% mtDNA copy number in muscle, 22% mtDNA copy number in blood. | | Mutations: | L304R, L304R | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 9, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Peripheral neuropathy exercise intolerance, easy fatigability, CPEO, abnormal EMG/NCV, ptosis, lactic acidosis, CPK abnormalities, short stature, FTT, abnormal VERS, developmental delay, abnormal respiratory enzymes, ragged red fibers. 37% mtDNA copy number in muscle, 20% mtDNA copy number in blood. | | Mutations: | L304R, L304R | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 12, Age of Death: n/a |
Back to top | Reference: | Tang et al, 2011; | | Description: | Peripheral neuropathy exercise intolerance, muscle weakness, easy fatigability, abnormal EMG/NCV, ptosis, CPK abnormalities, short stature, abnormal muscle histology, ragged red fibers, elevated pyruvate. 45% mtDNA copy number in blood. | | Mutations: | L304R, L304R | | Age group: | juvenile | | Age of Onset: n/a, Age of Patient: 23, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Peripheral neuropathy, exercise intolerance, muscle weakness, abnormal EMG/NCV, ptosis, lactic acidosis, COX deficiency, CPEO. 58% mtDNA copy number in blood. | | Mutations: | L304R, L304R | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 9, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Seizures, ptosis, elevated transaminases, lactic acidosis, elevated dopamine, abnormal MRI. 48% mtDNA copy number in muscle, 26% mtDNA copy number in liver. | | Mutations: | L304R, L304R | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 4, Age of Death: n/a |
| Reference: | Van Goethem et al, 2003a; | | Description: | sensory ataxic neuropathy, PEO, dysarthria. | | Mutations: | A467T, L304R | | Age group: | adult | | Age of Onset: 25, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Van Goethem et al, 2001; | | Description: | ocular symptoms, psychiatric symptoms, ptosis, opthalmoplegia, muscle weakness, distal sensory neuropathy, areflexia, pes cavus. | | Mutations: | A467T, L304R | | Age group: | adult | | Age of Onset: 25, Age of Patient: 38, Age of Death: n/a |
| Reference: | Van Goethem et al, 2001; | | Description: | ocular symptoms, ptosis, opthalmoplegia, dysphagia, muscle weakness, areflexia, retinopathy. | | Mutations: | A467T, L304R | | Age group: | adult | | Age of Onset: 24, Age of Patient: 34, Age of Death: n/a |
| Reference: | Van Goethem et al, 2001; | | Description: | psychiatric symptoms, ptosis, opthalmoplegia, dysphonia, dysphagia, muscle weakness, loss of weight, areflexia. | | Mutations: | A467T, L304R | | Age group: | juvenile | | Age of Onset: 16, Age of Patient: 28, Age of Death: n/a |
| Reference: | Bindu et al, 2016; | | Description: | CPEO, Sensory motor axonal neuropathy | | Mutations: | L304R | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 26, Age of Death: n/a |
| Reference: | Bindu et al, 2016; | | Description: | Mitochondrial spinocerebellar ataxia epilepsy syndrome, motor axonal neuropathy | | Mutations: | L304R | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 6, Age of Death: n/a |
Back to top | Reference: | Bindu et al, 2016; | | Description: | SANDO syndrome, Sensory motor demyelinating neuropathy | | Mutations: | L304R | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 10, Age of Death: n/a |
| Reference: | Horvath et al, 2006; | | Description: | PEO, Neuropathy. Affect sibling. | | Mutations: | L304R, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 60, Age of Patient: 68, Age of Death: n/a |
| Reference: | Horvath et al, 2006; | | Description: | PEO, myopathy, ataxia. | | Mutations: | L304R, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 45, Age of Patient: 74, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Cerebral cavernous malformation, teen onset CPEO. 110% mtDNA copy number in blood. | | Mutations: | A143V, L304R | | Age group: | juvenile | | Age of Onset: n/a, Age of Patient: 22, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Muscle weakness, ptosis, ophthalmoparesis/CPEO. 61% mtDNA copy number in blood. | | Mutations: | G737R, L304R | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 54, Age of Death: n/a |
Back to top | Reference: | Naimi et al, 2006; | | Description: | SCAE, age of onset 20, death at 27, ptosis and multiple deletions in muscle. | | Mutations: | L304R, W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: 20, Age of Patient: n/a, Age of Death: 27 |
| Reference: | Tang et al, 2011; | | Description: | Age 10, encephalopathy, ataxia, seizures, myoclonic seizures, pyramidal signs, dystonia, choria, muscle weakness, CPEO, ptosis, elevated alanine, abnormal EEG, MRS/lactate peak. 79% mtDNA copy number in blood. | | Mutations: | L304R, R1081P | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 10, Age of Death: n/a |
| Reference: | Navarro-Sastre et al, 2012; | | Description: | Alpers, psychomotor retardation, epileptic encephalopathy, progressive cerebral atrophy and altered liver enzymes, mtDNA depletion. Patient 9. Age of onset information obtained from corresponding author via email. | | Mutations: | G888D, L304R | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: 1.33 |
| Reference: | Navarro-Sastre et al, 2012; | | Description: | Alpers, intractable convulsions and severe epileptic status, high mtDNA depletion (8% residual mtDNA). Patient 10. Age of onset information obtained from corresponding author via email. | | Mutations: | L304R | | SNPs: | Y282D | | Age group: | infantile | | Age of Onset: 1.75, Age of Patient: n/a, Age of Death: n/a |
|
| G888D | | | | Number of patients: (with G888D) | 2 | | Non-allelic with: | W748S (50%) L304R (50%) |  | Show Patient Data |
| Patient data are sorted by mutation combination frequency. | Reference: | Tang et al, 2011; | | Description: | Seizures, ptosis, hepatic failure, ketosis, lactic acidosis, organic aciduria/tiglyglycine, elevated pyruvate, microcephaly, abnormal MRI. 33% mtDNA copy number in muscle, 46% mtDNA copy number in blood, ETC low. | | Mutations: | G888D, W748S | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 2, Age of Death: n/a |
| Reference: | Navarro-Sastre et al, 2012; | | Description: | Alpers, psychomotor retardation, epileptic encephalopathy, progressive cerebral atrophy and altered liver enzymes, mtDNA depletion. Patient 9. Age of onset information obtained from corresponding author via email. | | Mutations: | G888D, L304R | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: 1.33 |
|
|
|
|
|
The following information is based on existing patient data and pathogenic cluster assignment.
Pathogenicity information for a patient with mutations in Clusters 1 and 3: Age of onset information is extracted from a total of 18 patients and/ or patient families. | Age of onset | | |
18- 9- | 9
| 2
| 2
| 5
| | | infantile | childhd | juvenile | adult | | | 50% | 11% | 11% | 28% | |
All mutations mapping within the pathogenic clusters are at high risk for pathogenicity. In general, a patient must have a pathogenic mutation in both of his/ her POLG genes to develop a POLG-related syndrome.  | Symptoms described in patients with cluster3-cluster1 mutations | |
| Symptoms in patients with combination
cluster1:cluster3 | | Movement disorder (ataxia) | 44.4% | | Encephalopathy | 33.3% | | Developmental delay | 33.3% | | Epilepsy | 27.8% | | PEO | 27.8% | | Failure to thrive | 22.2% | | Hypotonic | 22.2% | | Peripheral neuropathy | 16.7% | | Ptosis | 16.7% | | Dysarthria | 16.7% | | Lactic acidosis | 11.1% | | Ragged red fibers | 11.1% | | Muscle weakness | 11.1% | | Liver failure | 11.1% | | Headache/ migraine | 11.1% | | Dementia | 11.1% | | Retardation | 11.1% | | GI dysmotility | 11.1% | | Myoclonic seizures | 5.6% | | Sensory ataxia | 5.6% | | Polyneuropathy | 5.6% | | Demyelinating neuropathy | 5.6% | | Axonal sensorimotor polyneuropathy | 5.6% | | Abnormal muscle ultrastructure | 5.6% | | Exercise intolerance | 5.6% | | Cox-negative | 5.6% | | Diplopia | 5.6% | | Liver dysfunction | 5.6% | | Growth retardation | 5.6% | | Vomiting | 5.6% | | GI reflux | 5.6% | | Cyclic vomiting | 5.6% | | Delayed gastric emptying | 5.6% | | Tremor | 5.6% | | Hearing loss | 5.6% |
| | Data gathered from clinical descriptions for 18 patients |
| Symptoms by group | | Developmental Delay | 50.0% | | Ataxia | 44.4% | | CPEO | 38.9% | | Seizures | 33.3% | | Neuropathy | 27.8% | | Alpers syndrome | 22.2% | | CNS symptoms | 22.2% | | Hypotonia | 22.2% | | GI symptoms | 16.7% | | Hepatopathy | 16.7% | | Myopathy | 16.7% | | Migraines | 11.1% | | Other | 5.6% |
| | [Show grouping information] |
|
|
|