Mutation Query
| | | | Allele 1: | G848S | | Allele 2: | R227W | Allelic information known | | Refine query |
|
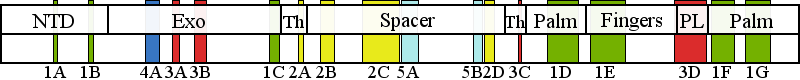
| | | Residue R227 | | Cluster assignment: | | | Cluster description: | Distal accessory subunit interface | | Subcluster: | 4A (residues 224-244) | | Subcluster description: | These mutations map to the exo domain along the distal accessory subunit interface. | | POLG domain: | Exonuclease domain |
| Residue G848 | | Cluster assignment: | | | Cluster description: | Polymerase active site and environs | | Subcluster: | 1D (residues 848-895) | | Subcluster description: | This subcluster forms a large portion of the pol active site and contains two highly conserved motifs that are found in all family A polymerases: the RR loop (motif 2) and motif A (Loh and Loeb, 2005). | | POLG domain: | Polymerase domain |
|
|
Mutation Information
|
| G848S | | | | Number of patients: (with G848S) | 66 | | Found together with: | |  | Show Patient Data |
| Patient data are sorted by mutation combination frequency. | Reference: | Ashley et al, 2008; | | Description: | Hepatopathy, Epilepsy, Hepatopathy, | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 1.5, Age of Patient: n/a, Age of Death: n/a |
| Reference: | de Vries et al, 2007; | | Description: | Onset at 6 months with FTT, dementia, hypotonia, seizures, hepatopathy, aplasia cutis, delayed myelinisation. Death at 15 months. | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: n/a, Age of Death: 1.3 |
| Reference: | Ferrari et al, 2005; | | Description: | He developed normally in the first 6 months. Refusal of food and failure to thrive were noted from the 7th month of life. At 1 year of age, increased transaminase levels in the blood were accompanied by liver enlargement and hyperecogenicity. Motor development was delayed. hypoglycaemic episodes and lactic acidosis. A liver biopsy at the age of 16 months revealed steatosis and fibrosis. At 20 months of age, he had numerous episodes of status epilepticus and epilepsia partialis continua. The patient died at 2.5 years of age during an episode of status epilepticus. Alpers’ syndrome. | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: n/a, Age of Death: 2.5 |
| Reference: | Kollberg et al, 2006; | | Description: | Ragged Red Fibers, COX-Deficient Fibers. The boy had feeding difficulties with failure to thrive since 3 months of age. Delayed motor development. general muscle weakness and ataxia. compromised liver function. | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 0.33, Age of Patient: n/a, Age of Death: 1.33 |
Back to top | Reference: | Stewart et al, 2009; | | Description: | Alpers, Epilepsy, developmental delay, COX deficient fibres. | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Stewart et al, 2009; | | Description: | Alpers, seizures, developmental delay, liver failure, hypotonia, impaired vision, renal stones, mtDNA depletion in muscle. 5% COX deficient fibers. | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 0.7, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Developmental delay, dementia, seizures, Alpers | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 2, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Developmental delay, dementia, seizures, Alpers, Ataxia | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 2, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Developmental delay, dementia, seizures, Alpers, Lactic acidosis, Cortical athrophy, hypoglycemia | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 1.5, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Wong et al, 2008; | | Description: | Developmental delay, dementia, seizures, Alpers, Lactic acidosis, Stroke | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 2, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Developmental delay, dementia, seizures, Alpers, Stroke, ataxia, exercise intolerance | | Mutations: | A467T, G848S | | Age group: | childhood | | Age of Onset: 9, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Developmental delay, dementia, seizures, Alpers, hypotonia, failure to thrive | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 0.8, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Nguyen et al, 2005; | | Description: | Refractory seizures, psychomotor regression, liver disease, Transient hypoglycemia, Transient lactic academia, Elevated CSF protein, presented with epilepsia partialis continua, | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 0.9, Age of Patient: n/a, Age of Death: 1.8 |
| Reference: | Nguyen et al, 2005; | | Description: | Refractory seizures, psychomotor regression, liver disease, presented with liver failure | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: 1.3 |
Back to top | Reference: | Saneto et al, 2010; | | Description: | Cyclic vomiting, episodic hypoglycemia then seizure onset at 6 years, generalized seizures, GI dysmotility, eyelid ptosis,ophthalmoplegia, sensorineural hearning loss, absent smooth pursuit eye movements | | Mutations: | A467T, G848S | | Age group: | childhood | | Age of Onset: 6, Age of Patient: 11, Age of Death: n/a |
| Reference: | Hunter et al, 2011; | | Description: | Myoclonic Seizures, Epilepsia partialis continua, Developmental Delay or Regression, ataxia, Abnormal Liver Enzymes, Serum Lactate, liver mtDNA depletion, Alpers | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: n/a, Age of Death: 3.33 |
| Reference: | Hunter et al, 2011; | | Description: | Myoclonic Seizures, Generalized tonic-clonic seizures, Focal motor seizures, Developmental Delay or Regression, hypotonia, tremor, Abnormal Liver Enzymes, Serum Lactate, liver mtDNA depletion, death by liver failure, Alpers | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 0.83, Age of Patient: n/a, Age of Death: 0.92 |
| Reference: | McCoy et al, 2011; | | Description: | focal seizures, elevated liver transaminases, Elevated CSF protein, patient died several weeks later | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 0.8, Age of Patient: n/a, Age of Death: 1 |
| Reference: | Scalais et al, 2012; | | Description: | presented with hypoglycemia, lactic acidosis, moderate ketosis and liver dysfunction, generalized hypotonia, progressive jaundice and abdominal distension with ascites, status epilepticus with generalized tonico-clonic seizures. She had an intermittent tremor, moderate truncal ataxia with a wide-based gait, myoclonic seizures, epilepsia partialis continua, | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 0.29, Age of Patient: n/a, Age of Death: 5 |
Back to top | Reference: | de Vries et al, 2008; | | Description: | Delayed motor and mental development, From the age of 3 years his psychomotor development declined and epilepsy, visual impairment and sensorineuronal deafness, epilepsia partialis continua. Bilateral ptosis, generalized hypotonia, ataxia, absent reflexes and hyperlaxity by age 5. | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 3, Age of Patient: 5, Age of Death: n/a |
| Reference: | Hasselmann et al, 2010; | | Description: | Alpers, status epilepticus and somnolence, ataxia with dysmetria and muscular hypotonia. Healthy unrelated parents. Laboratory investigations revealed myoclonic epilepsy with ragged red fibers, | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 2.2, Age of Patient: 3.5, Age of Death: 5.5 |
| Reference: | Tzoulis et al, 2013; | | Description: | Epilepsy, stroke-like episodes. | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 0.6, Age of Patient: n/a, Age of Death: 0.6 |
| Reference: | Roels et al, 2009; | | Description: | mtDNA depletion. severe hypoglycemia, liver biopsy shows micronodular cirrhosis. At 3 y 9 m, status epilepticus develops which occurs again at 4 y and is difficult to control. myoclonia. She dies from bronchopulmonary infection when 5 y old. COX negative fibers. | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 0.25, Age of Patient: 4, Age of Death: 5 |
| Reference: | Tzoulis et al, 2014; | | Description: | Epilepsy, stroke-like episode. | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 0.6, Age of Patient: n/a, Age of Death: 0.6 |
Back to top | Reference: | Ashley et al, 2008; | | Description: | Reported as Alpers, onset at 6 years, presenting encephalopathy with epilepsy, hepatopathy absent, and movement disorder (ataxia). | | Mutations: | G848S, W748S | | Age group: | childhood | | Age of Onset: 6, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Isohanni et al, 2011; | | Description: | Partial status epilepticus, epilepsia partialis, continua, hemiparesis, myoclonus, loss of vision, optical atrophy, pschomotor regression, liver dysfunction, death via pneumonia. | | Mutations: | G848S, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: 11 |
| Reference: | Taanman et al, 2009; | | Description: | Onset at 6.5 years with alpers syndrome and death at 7.8 years. 23% mtDNA copy number in liver. | | Mutations: | G848S, W748S | | SNPs: | E1143G | | Age group: | childhood | | Age of Onset: 6.5, Age of Patient: n/a, Age of Death: 7.8 |
| Reference: | Tang et al, 2011; | | Description: | Headaches/migrains, seizures, intractable seizure, cortical blindness. 32% mtDNA copy number in blood. | | Mutations: | G848S, W748S | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 7, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Hypotonia, myoclonus, or myoclonic seizures, intractable seizure, hepatic failure, abnormal EEG. 38% mtDNA copy number in blood. | | Mutations: | G848S, W748S | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 5, Age of Death: n/a |
Back to top | Reference: | Wong et al, 2008; | | Description: | developmental delay, dementia, seizures, Alpers, Family history of Alpers syndrome. | | Mutations: | G848S, Q497H, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Davidzon et al, 2005; | | Description: | hepatocerebral syndrome, alpers, developmental delay, seizures, and hepatopathy leading to liver failure, liver mtDNA depletion | | Mutations: | G848S, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 0.7, Age of Patient: n/a, Age of Death: 2.5 |
| Reference: | Davidzon et al, 2005; | | Description: | hepatocerebral syndrome, alpers, developmental delay, seizures, and hepatopathy leading to liver failure, liver mtDNA depletion, microvesicular steatosis, | | Mutations: | G848S, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: 6.5 |
| Reference: | Davidzon et al, 2005; | | Description: | hepatocerebral syndrome, alpers, developmental delay, seizures, and hepatopathy leading to liver failure, liver mtDNA depletion, microvesicular steatosis. | | Mutations: | G848S, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 0.1, Age of Patient: n/a, Age of Death: 0.9 |
| Reference: | Davidzon et al, 2005; | | Description: | hepatocerebral syndrome, alpers, developmental delay, seizures, and hepatopathy leading to liver failure, microvesicular steatosis, fibrosis | | Mutations: | G848S, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 0.3, Age of Patient: n/a, Age of Death: 1 |
Back to top | Reference: | Nguyen et al, 2005; | | Description: | Refractory seizures, psychomotor regression, liver disease, presenting symptom was status epilepticus, Transient hypoglycemia, Transient lactic acidemia | | Mutations: | G848S, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: 1.7 |
| Reference: | Wolf et al, 2009; | | Description: | Developemental delay, status epilepticus onset at 6 years 7 months, valproic acid therapy 6 weeks, liver failure with transplant, epilepsia partialis continua, elevated CSF lactate, elevated CSF protein, mtDNA depletion in liver | | Mutations: | G848S, W748S | | Age group: | infantile | | Age of Onset: 6.6, Age of Patient: 7.8, Age of Death: n/a |
| Reference: | Wolf et al, 2009; | | Description: | status epilepticus onset epilepsia partialis continua, elevated CSF lactate, elevated CSF protein, | | Mutations: | G848S, W748S | | Age group: | childhood | | Age of Onset: 10.67, Age of Patient: n/a, Age of Death: 11.5 |
| Reference: | Saneto et al, 2010; | | Description: | seizure onset at 2 years, complex partial seizure and epilepsia partialis continua myoclonus, Truncal ataxia, intention tremor | | Mutations: | G848S, W748S | | Age group: | infantile | | Age of Onset: 2, Age of Patient: 4, Age of Death: n/a |
| Reference: | Hunter et al, 2011; | | Description: | Myoclonic Seizures, Generalized tonic-clonic seizures, Focal motor seizures, Epilepsia partialis continua, Developmental Delay or Regression, ataxia, Abnormal Liver Enzymes, Serum Lactate, Alpers | | Mutations: | G848S, W748S | | Age group: | childhood | | Age of Onset: 6, Age of Patient: n/a, Age of Death: 6.75 |
Back to top | Reference: | Hunter et al, 2011; | | Description: | Myoclonic Seizures, Generalized tonic-clonic seizures, Focal motor seizures, Epilepsia partialis continua, Developmental Delay or Regression, ataxia, Abnormal Liver Enzymes, Serum Lactate, liver mtDNA depletion, Alpers | | Mutations: | G848S, W748S | | Age group: | infantile | | Age of Onset: 2, Age of Patient: n/a, Age of Death: 6.33 |
| Reference: | Brunetti-Pierri et al, 2008; | | Description: | He exhibited a very similar phenotype to his sister, with a catastrophic onset of intractable seizures at the age of 9 months, progressive encephalopathy with epilepsia partialis continua and myoclonic jerks, and developmental regression. MRI imaging was not performed. | | Mutations: | G848S, Q497H, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: 1.25, Age of Death: n/a |
| Reference: | Brunetti-Pierri et al, 2008; | | Description: | He exhibited a very similar phenotype to his sister, with a catastrophic onset of intractable seizures at the age of 9 months, progressive encephalopathy with epilepsia partialis continua and myoclonic jerks, and developmental regression. MRI imaging was not performed. | | Mutations: | G848S, Q497H, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: 1.25, Age of Death: n/a |
| Reference: | Brunetti-Pierri et al, 2008; | | Description: | The index patient presented at the age of 8 months with microcephaly, seizures, and developmental delay. From the time of the first observation she rapidly developed epilepsia partialis continua, multifocal myoclonic jerks, and progressive psychomotor retardation. MRI performed at 8 months was generally unremarkable. By 9 months of age the brain MRI showed signifi cant changes including a diffuse severe cerebral and cerebellar atrophy involving gray and white matter. | | Mutations: | G848S, Q497H, W748S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 0.6, Age of Death: 1 |
| Reference: | Lamantea et al, 2002; | | Description: | PEO. cytochrome c oxidase negative ragged red fibers, and multiple mtDNA deletions in skeletal muscle. | | Mutations: | G848S, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Tang et al, 2011; | | Description: | Ocular bulbar weakness, peripheral neuropathy, muscle weakness, CPEO, ptosis, hypothyroidism, abnormal muscle histology, abnormal muscle ultrastructure, large mitochondrial proliferation, ragged red fibers. 96% mtDNA copy number in blood. Patient II-44. | | Mutations: | G848S, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 81, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Alpers | | Mutations: | G848S, P587L | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Kollberg et al, 2005; | | Description: | PEO, severe ophthalmoplegia and ptosis, which appeared insidiously and had been present clinically since the age of 55. Muscle biopsy at age 68 revealed mitochondrial myopathy, as well as muscle inflammation with granulomas. multiple mtDNA deletions in muscle tissue. | | Mutations: | G848S, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 55, Age of Patient: 75, Age of Death: n/a |
| Reference: | Weiss and Saneto, 2010; | | Description: | progressively droopy eyelids, double vision, gait instability, proximal arm weakness, ragged red and ragged blue (succinate dehydrogenase–positive) fibers in about 3% and 6% of all muscle fibers, bilateral ptosis, diplopia, dysarthria, myopathy involving the facial muscles, sensory ataxic polyneuropathy | | Mutations: | G848S, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 80, Age of Death: n/a |
| Reference: | Rouzier et al, 2013; | | Description: | CPEO, multiple mtDNa deletions. | | Mutations: | G848S | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 45, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Tang et al, 2011; | | Description: | Ataxia, peripheral neuropathy, hearing loss, abnormal MRI, COX deficiency, developmental delay, seizure, delayed gastric emptying, CPK abnormalities, sister died at 2.5 years. 111% mtDNA copy number in blood. Patient II-49. | | Mutations: | A143V, G848S | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 38, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Evidence of maternal inheritance, developmental delay, hypotonia, dementia/encephalopathy, seizures, myoclonic seizures, hepatic failure, elevated transaminases, high CSF lactate, abnormal EEG, leukodystrophy, MRS/lactate peak, abnormal MRI. 71% mtDNA copy number in blood. | | Mutations: | A143V, G848S | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 4, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Leigh disease, developmental delay, FTT, abnormal EEG/MRI, hypotonia, seizures, vomiting, hemiplegia, behavior change. 136% mtDNA copy number in muscle and 57% mtDNA copy number in blood. | | Mutations: | A143V, G848S | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 4, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Seizures, intractable seizure, abnormal EEG, abnormal MRI. 43% mtDNA copy number in blood. | | Mutations: | A143V, G848S | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 8, Age of Death: n/a |
| Reference: | Giordano et al, 2009; | | Description: | Born with severe hypotonia, general muscle weakness, hearing loss, GI problems, mild cerbral atrophy, mtDNA depletion in muscle and GI, death via respiratory failure at 20 days. | | Mutations: | G848S, R227W | | Age group: | infantile | | Age of Onset: 0.01, Age of Patient: n/a, Age of Death: 0.1 |
Back to top | Reference: | Calvo et al, 2012; | | Description: | Complex I deficiency, Hypotonia, lethargy, respiratory distress, metabolic acidosis, FTT, encephalopathy | | Mutations: | G848S, R227W | | Age group: | infantile | | Age of Onset: 0.01, Age of Patient: 0.06, Age of Death: n/a |
| Reference: | Deschauer et al, 2007; | | Description: | vomiting, headache, sensory ataxia, areflexia, focal seizures, status epilepticus. MELAS. | | Mutations: | G848S, R627Q | | Age group: | adult | | Age of Onset: 21, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Schulte et al, 2009; | | Description: | At 35 years of age, dysarthria/dysphagia, sensory neuropathy, PEO, distal muscle wasting, phobia anxiety disorder, mild cerebellar atrophy (MRI) | | Mutations: | G848S, R627Q | | Age group: | adult | | Age of Onset: 29, Age of Patient: 35, Age of Death: n/a |
| Reference: | Stewart et al, 2009; | | Description: | Ataxia, PEO, axonal neuropathy, cerebellar atrophy. 3% COX deficient fibers. | | Mutations: | G746S, G848S | | SNPs: | E1143G | | Age group: | juvenile | | Age of Onset: 16, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Lax et al, 2012a; | | Description: | CPEO, peripheral neuropathy, ataxia, dysarthria, COX-deficient fibers, presence of mitochondrial dna deletions in muscle, Severe sensory and moderate motor neuronopathy, Distal and proximal neurogenic change | | Mutations: | G746S, G848S | | Age group: | adult | | Age of Onset: 26, Age of Patient: 36, Age of Death: n/a |
Back to top | Reference: | Kurt et al, 2010; | | Description: | pschomotor delay, seizures, liver disease, GI dysmotility, lactic acidosis. | | Mutations: | G848S, P1073L | | Age group: | infantile | | Age of Onset: 0.4, Age of Patient: n/a, Age of Death: 0.9 |
| Reference: | Tang et al, 2011; | | Description: | Hypotonia, FTT, gastro-oesophaeal reflux. 36% mtDNA copy number in muscle, 62% mtDNA copy number in blood. | | Mutations: | G848S, P1073L | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 2, Age of Death: n/a |
| Reference: | Lax et al, 2012b; | | Description: | Ataxia, Dementia, Peripheral neuropathy, Areflexia, Myopathy, fatigue, depression, Ophthalmoplegia, Dysarthria, Osteoporosis, arPEO. | | Mutations: | G848S, S1104C | | Age group: | adult | | Age of Onset: 22, Age of Patient: 59, Age of Death: n/a |
| Reference: | Betts-Henderson et al, 2009; | | Description: | The patient first presented at the age of 22 years with left-sided ptosis, which slowly progressed over the next 20 years to an almost complete ophthalmoplegia. He developed proximal muscle weakness at age 50 and his first Parkinsonian features at age 51. rigidity, tremor, bradykinesia and difficulty performing fine motor tasks. Proximal myopathy. sensorimotor neuropathy. Dysphagia. | | Mutations: | G848S, S1104C | | Age group: | adult | | Age of Onset: 22, Age of Patient: 57, Age of Death: 59 |
| Reference: | Taanman et al, 2009; | | Description: | Hypotonia, microcephaly, abnormal MRI, hepatomegaly, FTT, hypoglycaemia, ptosis. Onset at 6 months with Leighs syndrome and death at 23 months. 3% mtDNA copy number in muscle and 12% mtDNA copy number in liver. | | Mutations: | G848S, R232H | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: n/a, Age of Death: 2 |
Back to top | Reference: | Tang et al, 2011; | | Description: | Developmental delay, hypotonia, dementia/encephalopathy, exercise intolerance, muscle weakness, easy fatigability, ptosis, GI reflux, delayed gastric emptying, cyclic vomiting, hepatic failure, elevated transaminases, lactic acidosis, short statue, FTT. 53% mtDNA copy number in blood. | | Mutations: | G848S, R1096C | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 2, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Seizures, SIDS/unexplained death, MRS/lactate peak, abnormal MRI, abnormal respiratory enzymes. 7% mtDNA copy number in muscle. | | Mutations: | G848S, G848S | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 5, Age of Death: n/a |
|
| R227W | | | | Number of patients: (with R227W) | 7 | | Found together with: | |  | Show Patient Data |
| Patient data are sorted by mutation combination frequency. | Reference: | Agostino et al, 2003; | | Description: | PEO, bilateral ptosis, severe limita- tion of ocular motility, and a mosaic distribution of ragged-red and cytochrome c oxidase–negative fibers in the muscle biopsy. All patients had multiple deletions of muscle mtDNA. | | Mutations: | P587L, R227W | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 48, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Giordano et al, 2009; | | Description: | Born with severe hypotonia, general muscle weakness, hearing loss, GI problems, mild cerbral atrophy, mtDNA depletion in muscle and GI, death via respiratory failure at 20 days. | | Mutations: | G848S, R227W | | Age group: | infantile | | Age of Onset: 0.01, Age of Patient: n/a, Age of Death: 0.1 |
| Reference: | Calvo et al, 2012; | | Description: | Complex I deficiency, Hypotonia, lethargy, respiratory distress, metabolic acidosis, FTT, encephalopathy | | Mutations: | G848S, R227W | | Age group: | infantile | | Age of Onset: 0.01, Age of Patient: 0.06, Age of Death: n/a |
Back to top | Reference: | de Vries et al, 2007; | | Description: | Onset at 4 months with FTT, dementia, hypotonia, seizures, myoclonus, GI problems, hepatopathy, hearing loss, delayed myelinisation. Death at 10 months | | Mutations: | A467T, R227W | | Age group: | infantile | | Age of Onset: 0.3, Age of Patient: n/a, Age of Death: 0.8 |
| Reference: | de Vries et al, 2007; | | Description: | Severe childhood multi-system disorder- age of onset 4-7 months, age of death 26-43 months. Initial symptom was failure to thrive. Other symptoms included severe retardation, GI problems and hypotonia. Also hearing loss and elevated lactate in serum and CSF. Ragged red fibers were observed indicating mtDNA depletion | | Mutations: | D1184N, R227W | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: n/a, Age of Death: 2 |
|
|
|
|
|
The following information is based on existing patient data and pathogenic cluster assignment.
Pathogenicity information for a patient with mutations in Clusters 1 and 4: Age of onset information is extracted from a total of 5 patients and/ or patient families. | Age of onset | | |
5- 3- | 4
| 0
| 0
| 1
| | | infantile | childhd | juvenile | adult | | | 80% | 0% | 0% | 20% | |
All mutations mapping within the pathogenic clusters are at high risk for pathogenicity. In general, a patient must have a pathogenic mutation in both of his/ her POLG genes to develop a POLG-related syndrome.  | Symptoms described in patients with cluster1-cluster4 mutations | |
| Symptoms in patients with combination
cluster1:cluster4 | | Hypotonic | 80.0% | | Failure to thrive | 60.0% | | GI problems | 40.0% | | Hearing loss | 40.0% | | Ragged red fibers | 20.0% | | Muscle weakness | 20.0% | | Ptosis | 20.0% | | PEO | 20.0% | | Encephalopathy | 20.0% | | Retardation | 20.0% | | Respiratory deficiency | 20.0% | | Hepatomegaly | 20.0% | | Microcephaly | 20.0% |
| | Data gathered from clinical descriptions for 5 patients |
| Symptoms by group | | Hypotonia | 80.0% | | Developmental Delay | 60.0% | | Other | 60.0% | | CPEO | 40.0% | | GI symptoms | 40.0% | | Myopathy | 40.0% | | Hepatopathy | 20.0% |
| | [Show grouping information] |
|
|
|