Mutation Query
| | | | Allele 1: | P587L, T251I | | Allele 2: | P116Q | Allelic information known | | Refine query |
|
| | | Residue P116 | | Cluster assignment: | | | Cluster description: | Single Nucleotide Polymorphism | | POLG domain: | N-Terminal domain |
| Residue T251 | | Cluster assignment: | | | Cluster description: | Single Nucleotide Polymorphism | | POLG domain: | Exonuclease domain |
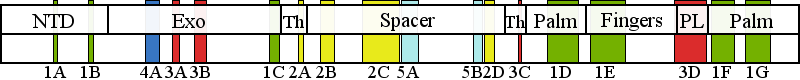
| Residue P587 | | Cluster assignment: | | | Cluster description: | Upstream DNA binding channel | | Subcluster: | 2C (residues 561-617) | | Subcluster description: | Subcluster 2C contains motif 1, which in Pol I was shown to fold into a loop that binds DNA in the channel (Loh and Loeb, 2005). Motif 1, together with subcluster 2D, form the major face of the putative DNA binding channel. | | POLG domain: | Spacer domain |
|
|
Mutation Information
|
| P587L | | | | Number of patients: (with P587L) | 53 | | Found together with: | PNF=Putatively Non-Functional enzyme |  | Show Patient Data |
| Patient data are sorted by mutation combination frequency. | Reference: | Agostino et al, 2003; | | Description: | PEO, bilateral ptosis, severe limita- tion of ocular motility, and a mosaic distribution of ragged-red and cytochrome c oxidase–negative fibers in the muscle biopsy. All patients had multiple deletions of muscle mtDNA. | | Mutations: | P587L, R227W | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 48, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Ashley et al, 2008; | | Description: | Alpers, epilepsy and hepatopathy, onset 5 months of age. | | Mutations: | P587L, R232G | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Blok et al, 2009; | | Description: | Exercise intolerance, CPEO, retinitis pigmentosa, diabetes, limb girdle weakness. 2nd patient with cataract and myopathy | | Mutations: | A467T, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 51, Age of Death: n/a |
Back to top | Reference: | Di Fonzo et al, 2003; | | Description: | PEO, axonal sensorimotor polyneuropathy | | Mutations: | P587L, R807P | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 60, Age of Patient: 71, Age of Death: n/a |
| Reference: | Di Fonzo et al, 2003; | | Description: | PEO, dysphagia | | Mutations: | P587L, R807P | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 45, Age of Patient: 52, Age of Death: n/a |
| Reference: | Ferreira et al, 2011; | | Description: | Ptosis, oculopharyngeal muscular dystrophy-like, pigmentary retinopathy. | | Mutations: | P587L, P648R | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 59, Age of Patient: 67, Age of Death: n/a |
| Reference: | Ferrari et al, 2005; | | Description: | Infantile hepatocerebral, presented at 3 months as hypotonia followed by mtDNA depletion in liver, dementia, and death at 6 months by ventilatory insufficiency. | | Mutations: | P587L, R232G | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.3, Age of Patient: n/a, Age of Death: 0.5 |
| Reference: | Gonzalez-Vioque et al, 2006; | | Description: | PEO with ptosis, mild atrial hypertrophy. Sister had PEO. | | Mutations: | M603L, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 61, Age of Patient: 65, Age of Death: n/a |
Back to top | Reference: | Horvath et al, 2006; | | Description: | PEO with myopathy | | Mutations: | P587L, P587L | | SNPs: | T251I, T251I | | Age group: | adult | | Age of Onset: 62, Age of Patient: 70, Age of Death: n/a |
| Reference: | Horvath et al, 2006; | | Description: | PEO with myopathy | | Mutations: | P587L, P587L | | SNPs: | T251I, T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 56, Age of Death: n/a |
| Reference: | Horvath et al, 2006; | | Description: | PEO with myopathy, chronic bronchitis. | | Mutations: | P587L, P587L | | SNPs: | T251I, T251I | | Age group: | adult | | Age of Onset: 63, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Horvath et al, 2006; | | Description: | PEO, Neuropathy. Affect sibling. | | Mutations: | L304R, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 60, Age of Patient: 68, Age of Death: n/a |
Back to top | Reference: | Horvath et al, 2006; | | Description: | PEO, myopathy, ataxia. | | Mutations: | L304R, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 45, Age of Patient: 74, Age of Death: n/a |
| Reference: | Lamantea et al, 2002; | | Description: | PEO. cytochrome c oxidase negative ragged red fibers, and multiple mtDNA deletions in skeletal muscle. | | Mutations: | G848S, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Lamantea et al, 2002; | | Description: | PEO. cytochrome c oxidase negative ragged red fibers, and multiple mtDNA deletions in skeletal muscle. | | Mutations: | P587L | | SNPs: | PNF, T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Lamantea et al, 2002; | | Description: | PEO. cytochrome c oxidase negative ragged red fibers, and multiple mtDNA deletions in skeletal muscle. | | Mutations: | P587L, R309L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Sarzi et al, 2007; | | Description: | Alpers, trunk hypotonia, seizures at 4 months, RC deficiency in liver. 28% mtDNA copy number in liver. | | Mutations: | C224Y, P587L | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.3, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Stewart et al, 2009; | | Description: | Mild bilateral ptosis, PEO. | | Mutations: | P587L, P587L | | SNPs: | T251I, T251I | | Age group: | adult | | Age of Onset: 41, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Stewart et al, 2009; | | Description: | PEO, ptosis, proximal weakness, multiple mtDNA deletions-LPCR. 25% COX-deficient fibers | | Mutations: | A467T, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 45, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Taanman et al, 2009; | | Description: | Vomiting, FTT, hypotonic, lactic acidemia, hypoglycemia, bilateral dense cataracts, hepatomegaly, jaundice, septicemia. infantile hepatocerebral mtDNA depletion. Mother with E1136K allele reported as asymptomatic. Father with T251I/P587L allele reported as asymptomatic. | | Mutations: | E1136K, P587L | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.01, Age of Patient: n/a, Age of Death: 0.5 |
| Reference: | Tang et al, 2011; | | Description: | N/A (as reported in Tang 2011 JMG) | | Mutations: | N1157S, P587L | | SNPs: | T251I | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 9, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Ocular bulbar weakness, peripheral neuropathy, muscle weakness, CPEO, ptosis, hypothyroidism, abnormal muscle histology, abnormal muscle ultrastructure, large mitochondrial proliferation, ragged red fibers. 96% mtDNA copy number in blood. Patient II-44. | | Mutations: | G848S, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 81, Age of Death: n/a |
Back to top | Reference: | Tang et al, 2011; | | Description: | Peripheral neuropathy, exercise intolerance, muscle cramps after exercise, easy fatigability, arrythmia, CPEO, abnormal EMG/NCV, ptosis, cataract, abnormal muscle histology, abnormal muscle ultrastructure, COX deficiency, ragged red fibers. 123% mtDNA copy number in blood. | | Mutations: | H932Y, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 31, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Peripheral neuropathy, ptosis, muscle weakness, CPK abnormalities. 77% mtDNA copy number in blood. | | Mutations: | H932Y, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 41, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Stroke/ischaemic episodes, peripheral neuropathy, CPEO, abnormal EMG/NCV, ptosis, muscle weakness, headache/migraine. 167% mtDNA copy number in blood. | | Mutations: | K1191N, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 39, Age of Death: n/a |
| Reference: | Van Goethem et al, 2003c; | | Description: | MNGIE-like, presented with GI symptoms, ptosis, neuropathy, muscle weakness. | | Mutations: | N864S, P587L | | SNPs: | T251I | | Age group: | juvenile | | Age of Onset: 15, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Alpers | | Mutations: | G848S, P587L | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Wong et al, 2008; | | Description: | Developmental delay, dementia, lactic acidosis, microcephaly, FTT, hearing loss, abnormal MRI. | | Mutations: | P587L, R853Q | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 0.2, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Developmental delay/dementia, liver dysfunction, lactic acidosis, fatigue, pancreatitis, cyclic vomiting. | | Mutations: | K1191R, P587L | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Kollberg et al, 2005; | | Description: | PEO, severe ophthalmoplegia and ptosis, which appeared insidiously and had been present clinically since the age of 55. Muscle biopsy at age 68 revealed mitochondrial myopathy, as well as muscle inflammation with granulomas. multiple mtDNA deletions in muscle tissue. | | Mutations: | G848S, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 55, Age of Patient: 75, Age of Death: n/a |
| Reference: | Burusnukul and de los Reyes, 2009; | | Description: | Global developmental delay, Central hypotonia, Torticollis, feeding difficulty at birth, myoclonic seizure, horizontal nystagmus, Intermittent estropia, Abnormal MRI | | Mutations: | P587L | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.4, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Lutz et al, 2009; | | Description: | Presented with feeding difficulties and hypotonia, At 4.5 months of age the patient contracted an influenza A infection. Concurrently she experienced hepatomegaly associated with severe hypoglycemia, Abnormalities in liver synthetic function, elevations in liver hepatocellular enzymes, elevated cerebrospinal fluid (CSF) protein, severely reduced mtDNA (3%) in liver. liver failure. She showed signs of ongoing respiratory difficulty, pancreatitis, and renal tubulopathy | | Mutations: | K1191R, P587L | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.166, Age of Patient: 0.416, Age of Death: 0.458 |
Back to top | Reference: | Weiss and Saneto, 2010; | | Description: | progressively droopy eyelids, double vision, gait instability, proximal arm weakness, ragged red and ragged blue (succinate dehydrogenase–positive) fibers in about 3% and 6% of all muscle fibers, bilateral ptosis, diplopia, dysarthria, myopathy involving the facial muscles, sensory ataxic polyneuropathy | | Mutations: | G848S, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 80, Age of Death: n/a |
| Reference: | Amiot et al, 2009; | | Description: | Peripheral neuropathy, PEO, ataxia, epilepsy, fatty liver | | Mutations: | D1184N, P587L | | SNPs: | T251I | | Age group: | childhood | | Age of Onset: 9, Age of Patient: n/a, Age of Death: 33 |
| Reference: | Amiot et al, 2009; | | Description: | Peripheral neuropathy, PEO, fatty liver | | Mutations: | D1184N, P587L | | SNPs: | T251I | | Age group: | childhood | | Age of Onset: 9, Age of Patient: n/a, Age of Death: 30 |
| Reference: | Aitken et al, 2009; | | Description: | Progressively blurred vision, diplopia, and longstanding bilateral ptosis (adPEO). She described occasional choking episodes after eating as well as fatigue and shortness of breath after minimal exertion. Lower limb examination revealed symmetric proximal limb weakness with reduced reflexes and flexor plantars. Tan- dem gait was hesitant. The initial differential diagnosis included Graves thyroid eye disease, a neuromuscular junction disor- der (myasthenia gravis or botulism), oculopharyngeal muscular dystrophy, Miller Fisher variant of Guillain-Barre syndrome, and progressive muscular dystrophy. Borderline myopathy. The muscle biopsy showed 13% cytochrome c ox- idase (COX)–deficient fibers, significant numbers of ragged red fibers but no excess of lipid or glycogen accumulation, and subsarcolemmal accumulation of abnormal mitochondria suggestive of a mitochondrial cytopathy. The patient presented 10 months later with an un- pleasant “jumping†sensation in her feet when at rest which was relieved by movement. Symptoms were worse at night and she also described sudden involuntary movements of her lower limbs (restless legs syndrome, RLS). | | Mutations: | P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 53, Age of Patient: 58, Age of Death: n/a |
| Reference: | Hanisch et al, 2014; | | Description: | Pareses, ptosis, ataxia, sensory neoropathy, motor neuropathy, axonal neuropathy, demyelinating neuropathy. | | Mutations: | P587L, R869Q | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 29, Age of Patient: 34, Age of Death: n/a |
Back to top | Reference: | Harris et al, 2010; | | Description: | Progressive ataxia, asymmetrical right ophthalmoplegia and encephalopathy. Seizures. Loss of extraocular movement on the right and right arm and leg spasticity and hyperreflexia. Elevated lactate level, a markedly depressed N-acetylaspartate level, and a markedly elevated choline level. Steroid-induced hyperglycemia. Progressive acute disseminated encephalomyelitis (ADEM). | | Mutations: | P587L | | SNPs: | T251I | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 4, Age of Death: 4.75 |
| Reference: | Lovan et al, 2013; | | Description: | diabetes mellitus 2, congestive heart failure, hypothyroidism, hypertension, depression with psychotic features and gastric bypass surgery. gait difficulties, cognitive impairment, ophthalmoplegia, resting tremor and peripheral neuropathy. Shooting pains and numbness in her lower extremities, occasional rectal incontinence. Presyncopal dizziness. Complicated diabetic retinopathy. sensory ataxia secondary to a sensorimotor polyneuropathy, chronic supranuclear ophthalmoplegia, and a resting tremor of her right hand. bilateral ptosis and marked temporal muscle wasting. Her sensory examination found loss of pinprick sensation, temperature and vibration bilaterally up to her hips. She had bilateral vibration loss at her fingertips. She had loss of proprioception at her toes. She had bilateral dysdiadochokinesia with significant ataxia on finger to nose testing. Arreflexic in her lower extremities. On cognitive testing she was found to have moderate dementia with significant deficits in registration and construction as well as ideomotor apraxia. | | Mutations: | A467T, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 55, Age of Patient: 60, Age of Death: n/a |
| Reference: | Scuderi et al, 2015; | | Description: | He presented migraine by age 7 and complained of fatigue during exercise by age 12. mild diffuse muscle hypotonia and flat feet. ptosis at 14. Brain MRI showed mild white matter hyperintensity. | | Mutations: | P587L | | SNPs: | P116Q, T251I | | Age group: | childhood | | Age of Onset: 7, Age of Patient: 14, Age of Death: n/a |
| Reference: | Scuderi et al, 2015; | | Description: | Presented deafness from childhood. migraine and fatigue. On neurological examination she had mild diffuse muscle weakness. Audiometric examination disclosed neurosensorial hypoacusia. mild anxiety with specific phobias concerning indoor environment and crowd. | | Mutations: | P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 29, Age of Patient: 37, Age of Death: n/a |
| Reference: | Scuderi et al, 2015; | | Description: | delayed psychomotor development. At age 3, neurological examination showed muscular hypotonia, joint laxity, absent deep tendon reflexes, broadbased gait, scapular winging, accentuation of lumbar lordosis and flat feet. intellectual disability. progressive cognitive impairment. mild myopathy. | | Mutations: | P587L | | SNPs: | P116Q, T251I | | Age group: | childhood | | Age of Onset: 3, Age of Patient: 8, Age of Death: n/a |
Back to top | Reference: | Scuderi et al, 2015; | | Description: | leg pain after physical activity. mild diffuse muscle hypotonia, hypotrophy, mild diffuse muscle weakness, joint laxity, scapular winging and increase of lumbar lordosis. Borderline intellectual functioning. | | Mutations: | P587L | | SNPs: | T251I | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 10, Age of Death: n/a |
| Reference: | Stewart et al, 2011; | | Description: | Alpers. nocturnal nausea followed by loss of consciousness without motor relinquishing. obsessive–compulsive disorder. | | Mutations: | P587L, P587L | | SNPs: | T251I, T251I | | Age group: | juvenile | | Age of Onset: 16, Age of Patient: 26, Age of Death: n/a |
| Reference: | Del Bo et al, 2003; | | Description: | PEO, complicated by dysphagia, myopathy. | | Mutations: | P587L | | SNPs: | PNF, T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 52, Age of Death: n/a |
| Reference: | Del Bo et al, 2003; | | Description: | PEO, complicated by dysphagia, myopathy. | | Mutations: | P587L, R807P | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 71, Age of Death: n/a |
| Reference: | Miguel et al, 2014; | | Description: | slowly progressive bilateral blepharoptosis, severe PEO, dysphagia, facial diparesis and exercise intolerance, presented, at the age of 60, with left-dominant postural tremor, reduced left arm swing during gait, postural instability, axonal sensory polyneuropathy, CT showed diffuse cerebral atrophy. Parkinsons. | | Mutations: | P587L | | SNPs: | PNF, T251I | | Age group: | adult | | Age of Onset: 46, Age of Patient: 66, Age of Death: n/a |
Back to top | Reference: | Ashley et al, 2008; | | Description: | Encephalopathy onset 16 years of age, presented with epilepsy. | | Mutations: | P587L, P589L, W748S | | Age group: | juvenile | | Age of Onset: 16, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Tzoulis et al, 2009; | | Description: | PEO, ptosis, muscle fatigue, diplopia, a mild external ophthalmoplegia affecting horizontal, but not vertical gaze. Tendonreflexes were diminished in the lower extremities and a distal sensory defect in stocking distribution was seen. | | Mutations: | P587L, W748S | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 45, Age of Patient: 50, Age of Death: n/a |
| Reference: | Ylönen et al, 2013; | | Description: | Akinetic-rigid Parkinsons. impaired balance, freezing, and increased salivation. Constipation and muscle cramps. He had arterial hypertension and mild normocytic anemia. cytochrome c oxidase-negative muscle fibers. | | Mutations: | P587L, W748S | | SNPs: | E1142G, T251I | | Age group: | adult | | Age of Onset: 49, Age of Patient: 65, Age of Death: n/a |
| Reference: | Horvath et al, 2006; | | Description: | PEO with myopathy | | Mutations: | P587L, V1106I | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 35, Age of Death: n/a |
|
| T251I | | | | Number of patients: (with T251I) | 53 | | Found together with: | PNF=Putatively Non-Functional enzyme |  | Show Patient Data |
| Patient data are sorted by mutation combination frequency. | Reference: | Blok et al, 2009; | | Description: | Exercise intolerance, CPEO, retinitis pigmentosa, diabetes, limb girdle weakness. 2nd patient with cataract and myopathy | | Mutations: | A467T, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 51, Age of Death: n/a |
| Reference: | Gonzalez-Vioque et al, 2006; | | Description: | PEO with ptosis, mild atrial hypertrophy. Sister had PEO. | | Mutations: | M603L, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 61, Age of Patient: 65, Age of Death: n/a |
| Reference: | Horvath et al, 2006; | | Description: | PEO, Neuropathy. Affect sibling. | | Mutations: | L304R, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 60, Age of Patient: 68, Age of Death: n/a |
| Reference: | Horvath et al, 2006; | | Description: | PEO, myopathy, ataxia. | | Mutations: | L304R, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 45, Age of Patient: 74, Age of Death: n/a |
Back to top | Reference: | Lamantea et al, 2002; | | Description: | PEO. cytochrome c oxidase negative ragged red fibers, and multiple mtDNA deletions in skeletal muscle. | | Mutations: | G848S, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Sarzi et al, 2007; | | Description: | Alpers, trunk hypotonia, seizures at 4 months, RC deficiency in liver. 28% mtDNA copy number in liver. | | Mutations: | C224Y, P587L | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.3, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Stewart et al, 2009; | | Description: | PEO, ptosis, proximal weakness, multiple mtDNA deletions-LPCR. 25% COX-deficient fibers | | Mutations: | A467T, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 45, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Taanman et al, 2009; | | Description: | Vomiting, FTT, hypotonic, lactic acidemia, hypoglycemia, bilateral dense cataracts, hepatomegaly, jaundice, septicemia. infantile hepatocerebral mtDNA depletion. Mother with E1136K allele reported as asymptomatic. Father with T251I/P587L allele reported as asymptomatic. | | Mutations: | E1136K, P587L | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.01, Age of Patient: n/a, Age of Death: 0.5 |
| Reference: | Tang et al, 2011; | | Description: | N/A (as reported in Tang 2011 JMG) | | Mutations: | N1157S, P587L | | SNPs: | T251I | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 9, Age of Death: n/a |
Back to top | Reference: | Tang et al, 2011; | | Description: | Ocular bulbar weakness, peripheral neuropathy, muscle weakness, CPEO, ptosis, hypothyroidism, abnormal muscle histology, abnormal muscle ultrastructure, large mitochondrial proliferation, ragged red fibers. 96% mtDNA copy number in blood. Patient II-44. | | Mutations: | G848S, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 81, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Peripheral neuropathy, exercise intolerance, muscle cramps after exercise, easy fatigability, arrythmia, CPEO, abnormal EMG/NCV, ptosis, cataract, abnormal muscle histology, abnormal muscle ultrastructure, COX deficiency, ragged red fibers. 123% mtDNA copy number in blood. | | Mutations: | H932Y, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 31, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Peripheral neuropathy, ptosis, muscle weakness, CPK abnormalities. 77% mtDNA copy number in blood. | | Mutations: | H932Y, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 41, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Stroke/ischaemic episodes, peripheral neuropathy, CPEO, abnormal EMG/NCV, ptosis, muscle weakness, headache/migraine. 167% mtDNA copy number in blood. | | Mutations: | K1191N, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 39, Age of Death: n/a |
| Reference: | Van Goethem et al, 2003c; | | Description: | MNGIE-like, presented with GI symptoms, ptosis, neuropathy, muscle weakness. | | Mutations: | N864S, P587L | | SNPs: | T251I | | Age group: | juvenile | | Age of Onset: 15, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Wong et al, 2008; | | Description: | Alpers | | Mutations: | G848S, P587L | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Developmental delay/dementia, liver dysfunction, lactic acidosis, fatigue, pancreatitis, cyclic vomiting. | | Mutations: | K1191R, P587L | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Kollberg et al, 2005; | | Description: | PEO, severe ophthalmoplegia and ptosis, which appeared insidiously and had been present clinically since the age of 55. Muscle biopsy at age 68 revealed mitochondrial myopathy, as well as muscle inflammation with granulomas. multiple mtDNA deletions in muscle tissue. | | Mutations: | G848S, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 55, Age of Patient: 75, Age of Death: n/a |
| Reference: | Burusnukul and de los Reyes, 2009; | | Description: | Global developmental delay, Central hypotonia, Torticollis, feeding difficulty at birth, myoclonic seizure, horizontal nystagmus, Intermittent estropia, Abnormal MRI | | Mutations: | P587L | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.4, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Lutz et al, 2009; | | Description: | Presented with feeding difficulties and hypotonia, At 4.5 months of age the patient contracted an influenza A infection. Concurrently she experienced hepatomegaly associated with severe hypoglycemia, Abnormalities in liver synthetic function, elevations in liver hepatocellular enzymes, elevated cerebrospinal fluid (CSF) protein, severely reduced mtDNA (3%) in liver. liver failure. She showed signs of ongoing respiratory difficulty, pancreatitis, and renal tubulopathy | | Mutations: | K1191R, P587L | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.166, Age of Patient: 0.416, Age of Death: 0.458 |
Back to top | Reference: | Weiss and Saneto, 2010; | | Description: | progressively droopy eyelids, double vision, gait instability, proximal arm weakness, ragged red and ragged blue (succinate dehydrogenase–positive) fibers in about 3% and 6% of all muscle fibers, bilateral ptosis, diplopia, dysarthria, myopathy involving the facial muscles, sensory ataxic polyneuropathy | | Mutations: | G848S, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 80, Age of Death: n/a |
| Reference: | Amiot et al, 2009; | | Description: | Peripheral neuropathy, PEO, ataxia, epilepsy, fatty liver | | Mutations: | D1184N, P587L | | SNPs: | T251I | | Age group: | childhood | | Age of Onset: 9, Age of Patient: n/a, Age of Death: 33 |
| Reference: | Amiot et al, 2009; | | Description: | Peripheral neuropathy, PEO, fatty liver | | Mutations: | D1184N, P587L | | SNPs: | T251I | | Age group: | childhood | | Age of Onset: 9, Age of Patient: n/a, Age of Death: 30 |
| Reference: | Aitken et al, 2009; | | Description: | Progressively blurred vision, diplopia, and longstanding bilateral ptosis (adPEO). She described occasional choking episodes after eating as well as fatigue and shortness of breath after minimal exertion. Lower limb examination revealed symmetric proximal limb weakness with reduced reflexes and flexor plantars. Tan- dem gait was hesitant. The initial differential diagnosis included Graves thyroid eye disease, a neuromuscular junction disor- der (myasthenia gravis or botulism), oculopharyngeal muscular dystrophy, Miller Fisher variant of Guillain-Barre syndrome, and progressive muscular dystrophy. Borderline myopathy. The muscle biopsy showed 13% cytochrome c ox- idase (COX)–deficient fibers, significant numbers of ragged red fibers but no excess of lipid or glycogen accumulation, and subsarcolemmal accumulation of abnormal mitochondria suggestive of a mitochondrial cytopathy. The patient presented 10 months later with an un- pleasant “jumping†sensation in her feet when at rest which was relieved by movement. Symptoms were worse at night and she also described sudden involuntary movements of her lower limbs (restless legs syndrome, RLS). | | Mutations: | P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 53, Age of Patient: 58, Age of Death: n/a |
| Reference: | Harris et al, 2010; | | Description: | Progressive ataxia, asymmetrical right ophthalmoplegia and encephalopathy. Seizures. Loss of extraocular movement on the right and right arm and leg spasticity and hyperreflexia. Elevated lactate level, a markedly depressed N-acetylaspartate level, and a markedly elevated choline level. Steroid-induced hyperglycemia. Progressive acute disseminated encephalomyelitis (ADEM). | | Mutations: | P587L | | SNPs: | T251I | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 4, Age of Death: 4.75 |
Back to top | Reference: | Lovan et al, 2013; | | Description: | diabetes mellitus 2, congestive heart failure, hypothyroidism, hypertension, depression with psychotic features and gastric bypass surgery. gait difficulties, cognitive impairment, ophthalmoplegia, resting tremor and peripheral neuropathy. Shooting pains and numbness in her lower extremities, occasional rectal incontinence. Presyncopal dizziness. Complicated diabetic retinopathy. sensory ataxia secondary to a sensorimotor polyneuropathy, chronic supranuclear ophthalmoplegia, and a resting tremor of her right hand. bilateral ptosis and marked temporal muscle wasting. Her sensory examination found loss of pinprick sensation, temperature and vibration bilaterally up to her hips. She had bilateral vibration loss at her fingertips. She had loss of proprioception at her toes. She had bilateral dysdiadochokinesia with significant ataxia on finger to nose testing. Arreflexic in her lower extremities. On cognitive testing she was found to have moderate dementia with significant deficits in registration and construction as well as ideomotor apraxia. | | Mutations: | A467T, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 55, Age of Patient: 60, Age of Death: n/a |
| Reference: | Scuderi et al, 2015; | | Description: | He presented migraine by age 7 and complained of fatigue during exercise by age 12. mild diffuse muscle hypotonia and flat feet. ptosis at 14. Brain MRI showed mild white matter hyperintensity. | | Mutations: | P587L | | SNPs: | P116Q, T251I | | Age group: | childhood | | Age of Onset: 7, Age of Patient: 14, Age of Death: n/a |
| Reference: | Scuderi et al, 2015; | | Description: | Presented deafness from childhood. migraine and fatigue. On neurological examination she had mild diffuse muscle weakness. Audiometric examination disclosed neurosensorial hypoacusia. mild anxiety with specific phobias concerning indoor environment and crowd. | | Mutations: | P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 29, Age of Patient: 37, Age of Death: n/a |
| Reference: | Scuderi et al, 2015; | | Description: | delayed psychomotor development. At age 3, neurological examination showed muscular hypotonia, joint laxity, absent deep tendon reflexes, broadbased gait, scapular winging, accentuation of lumbar lordosis and flat feet. intellectual disability. progressive cognitive impairment. mild myopathy. | | Mutations: | P587L | | SNPs: | P116Q, T251I | | Age group: | childhood | | Age of Onset: 3, Age of Patient: 8, Age of Death: n/a |
| Reference: | Scuderi et al, 2015; | | Description: | leg pain after physical activity. mild diffuse muscle hypotonia, hypotrophy, mild diffuse muscle weakness, joint laxity, scapular winging and increase of lumbar lordosis. Borderline intellectual functioning. | | Mutations: | P587L | | SNPs: | T251I | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 10, Age of Death: n/a |
Back to top | Reference: | Rouzier et al, 2013; | | Description: | CPEO, multiple mtDNa deletions. | | Mutations: | G848S | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 45, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Horvath et al, 2006; | | Description: | PEO with myopathy | | Mutations: | P587L, P587L | | SNPs: | T251I, T251I | | Age group: | adult | | Age of Onset: 62, Age of Patient: 70, Age of Death: n/a |
| Reference: | Horvath et al, 2006; | | Description: | PEO with myopathy | | Mutations: | P587L, P587L | | SNPs: | T251I, T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 56, Age of Death: n/a |
| Reference: | Horvath et al, 2006; | | Description: | PEO with myopathy, chronic bronchitis. | | Mutations: | P587L, P587L | | SNPs: | T251I, T251I | | Age group: | adult | | Age of Onset: 63, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Stewart et al, 2009; | | Description: | Mild bilateral ptosis, PEO. | | Mutations: | P587L, P587L | | SNPs: | T251I, T251I | | Age group: | adult | | Age of Onset: 41, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Stewart et al, 2011; | | Description: | Alpers. nocturnal nausea followed by loss of consciousness without motor relinquishing. obsessive–compulsive disorder. | | Mutations: | P587L, P587L | | SNPs: | T251I, T251I | | Age group: | juvenile | | Age of Onset: 16, Age of Patient: 26, Age of Death: n/a |
| Reference: | Di Fonzo et al, 2003; | | Description: | PEO, axonal sensorimotor polyneuropathy | | Mutations: | P587L, R807P | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 60, Age of Patient: 71, Age of Death: n/a |
| Reference: | Di Fonzo et al, 2003; | | Description: | PEO, dysphagia | | Mutations: | P587L, R807P | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 45, Age of Patient: 52, Age of Death: n/a |
| Reference: | Del Bo et al, 2003; | | Description: | PEO, complicated by dysphagia, myopathy. | | Mutations: | P587L, R807P | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 71, Age of Death: n/a |
Back to top | Reference: | Lamantea et al, 2002; | | Description: | PEO. cytochrome c oxidase negative ragged red fibers, and multiple mtDNA deletions in skeletal muscle. | | Mutations: | P587L | | SNPs: | PNF, T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Del Bo et al, 2003; | | Description: | PEO, complicated by dysphagia, myopathy. | | Mutations: | P587L | | SNPs: | PNF, T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 52, Age of Death: n/a |
| Reference: | Miguel et al, 2014; | | Description: | slowly progressive bilateral blepharoptosis, severe PEO, dysphagia, facial diparesis and exercise intolerance, presented, at the age of 60, with left-dominant postural tremor, reduced left arm swing during gait, postural instability, axonal sensory polyneuropathy, CT showed diffuse cerebral atrophy. Parkinsons. | | Mutations: | P587L | | SNPs: | PNF, T251I | | Age group: | adult | | Age of Onset: 46, Age of Patient: 66, Age of Death: n/a |
| Reference: | Agostino et al, 2003; | | Description: | PEO, bilateral ptosis, severe limita- tion of ocular motility, and a mosaic distribution of ragged-red and cytochrome c oxidase–negative fibers in the muscle biopsy. All patients had multiple deletions of muscle mtDNA. | | Mutations: | P587L, R227W | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 48, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Ashley et al, 2008; | | Description: | Alpers, epilepsy and hepatopathy, onset 5 months of age. | | Mutations: | P587L, R232G | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Ferrari et al, 2005; | | Description: | Infantile hepatocerebral, presented at 3 months as hypotonia followed by mtDNA depletion in liver, dementia, and death at 6 months by ventilatory insufficiency. | | Mutations: | P587L, R232G | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.3, Age of Patient: n/a, Age of Death: 0.5 |
| Reference: | Tzoulis et al, 2009; | | Description: | PEO, ptosis, muscle fatigue, diplopia, a mild external ophthalmoplegia affecting horizontal, but not vertical gaze. Tendonreflexes were diminished in the lower extremities and a distal sensory defect in stocking distribution was seen. | | Mutations: | P587L, W748S | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 45, Age of Patient: 50, Age of Death: n/a |
| Reference: | Ylönen et al, 2013; | | Description: | Akinetic-rigid Parkinsons. impaired balance, freezing, and increased salivation. Constipation and muscle cramps. He had arterial hypertension and mild normocytic anemia. cytochrome c oxidase-negative muscle fibers. | | Mutations: | P587L, W748S | | SNPs: | E1142G, T251I | | Age group: | adult | | Age of Onset: 49, Age of Patient: 65, Age of Death: n/a |
| Reference: | Horvath et al, 2006; | | Description: | PEO with myopathy | | Mutations: | P587L, V1106I | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 35, Age of Death: n/a |
Back to top | Reference: | Ferreira et al, 2011; | | Description: | Ptosis, oculopharyngeal muscular dystrophy-like, pigmentary retinopathy. | | Mutations: | P587L, P648R | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 59, Age of Patient: 67, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Developmental delay, dementia, lactic acidosis, microcephaly, FTT, hearing loss, abnormal MRI. | | Mutations: | P587L, R853Q | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 0.2, Age of Death: n/a |
| Reference: | Hanisch et al, 2014; | | Description: | Pareses, ptosis, ataxia, sensory neoropathy, motor neuropathy, axonal neuropathy, demyelinating neuropathy. | | Mutations: | P587L, R869Q | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 29, Age of Patient: 34, Age of Death: n/a |
| Reference: | Lamantea et al, 2002; | | Description: | PEO. cytochrome c oxidase negative ragged red fibers, and multiple mtDNA deletions in skeletal muscle. | | Mutations: | P587L, R309L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: n/a, Age of Death: n/a |
|
| P116Q | | | | Number of patients: (with P116Q) | 3 | | Found as the only mutation: | 33% of entries (1 patient) | | Non-allelic with: | T251I (67%) P587L (67%) |  | Show Patient Data |
| Patient data are sorted by mutation combination frequency. | Reference: | Scuderi et al, 2015; | | Description: | He presented migraine by age 7 and complained of fatigue during exercise by age 12. mild diffuse muscle hypotonia and flat feet. ptosis at 14. Brain MRI showed mild white matter hyperintensity. | | Mutations: | P587L | | SNPs: | P116Q, T251I | | Age group: | childhood | | Age of Onset: 7, Age of Patient: 14, Age of Death: n/a |
| Reference: | Scuderi et al, 2015; | | Description: | delayed psychomotor development. At age 3, neurological examination showed muscular hypotonia, joint laxity, absent deep tendon reflexes, broadbased gait, scapular winging, accentuation of lumbar lordosis and flat feet. intellectual disability. progressive cognitive impairment. mild myopathy. | | Mutations: | P587L | | SNPs: | P116Q, T251I | | Age group: | childhood | | Age of Onset: 3, Age of Patient: 8, Age of Death: n/a |
| Reference: | Scuderi et al, 2015; | | Description: | By age 38 he complained fatigue. At the age of 40 he presented diabetes mellitus. Deep tendon reflexes were absent on neurological examination. | | Mutations: | | | SNPs: | P116Q | | Age group: | adult | | Age of Onset: 38, Age of Patient: 42, Age of Death: n/a |
|
|
|
|
The following information is based on PON-P2 mutation pathogenicity prediction software results. Cluster 2 mutation with a non-cluster-mapping mutation (SNP) P116Q Mutation pathogenicity prediction for mutation P116Q is unreliable. Mutation P116Q is outside of assigned pathogenic clusters, and therefore risk that it contributes to a POLG-related syndrome is low. See further details for residue 116. All mutations mapping into the pathogenic clusters are in high risk of being pathogenic. As a rule, a patient must have a pathogenic mutation in both of his/ her POLG genes to develop a POLG-related syndrome. |
|