Mutation Query
| | | | Allele 1: | P587L, T251I | | Allele 2: | A467T | Allelic information known | | Refine query |
|
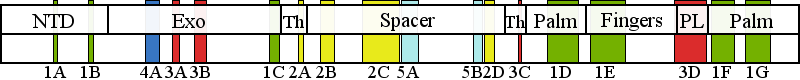
| | | Residue T251 | | Cluster assignment: | | | Cluster description: | Single Nucleotide Polymorphism | | POLG domain: | Exonuclease domain |
| Residue A467 | | Cluster assignment: | | | Cluster description: | Upstream DNA binding channel | | Subcluster: | 2A (residues 463-468) | | Subcluster description: | Subcluster 2A maps to a region of the thumb subdomain of the pol domain at the accessory subunit interface where A467T, N468D and L463F are positioned. | | POLG domain: | Polymerase domain |
| Residue P587 | | Cluster assignment: | | | Cluster description: | Upstream DNA binding channel | | Subcluster: | 2C (residues 561-617) | | Subcluster description: | Subcluster 2C contains motif 1, which in Pol I was shown to fold into a loop that binds DNA in the channel (Loh and Loeb, 2005). Motif 1, together with subcluster 2D, form the major face of the putative DNA binding channel. | | POLG domain: | Spacer domain |
|
|
Mutation Information
|
| P587L | | | | Number of patients: (with P587L) | 53 | | Found together with: | PNF=Putatively Non-Functional enzyme |  | Show Patient Data |
| Patient data are sorted by mutation combination frequency. | Reference: | Agostino et al, 2003; | | Description: | PEO, bilateral ptosis, severe limita- tion of ocular motility, and a mosaic distribution of ragged-red and cytochrome c oxidase–negative fibers in the muscle biopsy. All patients had multiple deletions of muscle mtDNA. | | Mutations: | P587L, R227W | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 48, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Ashley et al, 2008; | | Description: | Alpers, epilepsy and hepatopathy, onset 5 months of age. | | Mutations: | P587L, R232G | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Blok et al, 2009; | | Description: | Exercise intolerance, CPEO, retinitis pigmentosa, diabetes, limb girdle weakness. 2nd patient with cataract and myopathy | | Mutations: | A467T, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 51, Age of Death: n/a |
Back to top | Reference: | Di Fonzo et al, 2003; | | Description: | PEO, axonal sensorimotor polyneuropathy | | Mutations: | P587L, R807P | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 60, Age of Patient: 71, Age of Death: n/a |
| Reference: | Di Fonzo et al, 2003; | | Description: | PEO, dysphagia | | Mutations: | P587L, R807P | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 45, Age of Patient: 52, Age of Death: n/a |
| Reference: | Ferreira et al, 2011; | | Description: | Ptosis, oculopharyngeal muscular dystrophy-like, pigmentary retinopathy. | | Mutations: | P587L, P648R | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 59, Age of Patient: 67, Age of Death: n/a |
| Reference: | Ferrari et al, 2005; | | Description: | Infantile hepatocerebral, presented at 3 months as hypotonia followed by mtDNA depletion in liver, dementia, and death at 6 months by ventilatory insufficiency. | | Mutations: | P587L, R232G | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.3, Age of Patient: n/a, Age of Death: 0.5 |
| Reference: | Gonzalez-Vioque et al, 2006; | | Description: | PEO with ptosis, mild atrial hypertrophy. Sister had PEO. | | Mutations: | M603L, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 61, Age of Patient: 65, Age of Death: n/a |
Back to top | Reference: | Horvath et al, 2006; | | Description: | PEO with myopathy | | Mutations: | P587L, P587L | | SNPs: | T251I, T251I | | Age group: | adult | | Age of Onset: 62, Age of Patient: 70, Age of Death: n/a |
| Reference: | Horvath et al, 2006; | | Description: | PEO with myopathy | | Mutations: | P587L, P587L | | SNPs: | T251I, T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 56, Age of Death: n/a |
| Reference: | Horvath et al, 2006; | | Description: | PEO with myopathy, chronic bronchitis. | | Mutations: | P587L, P587L | | SNPs: | T251I, T251I | | Age group: | adult | | Age of Onset: 63, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Horvath et al, 2006; | | Description: | PEO, Neuropathy. Affect sibling. | | Mutations: | L304R, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 60, Age of Patient: 68, Age of Death: n/a |
Back to top | Reference: | Horvath et al, 2006; | | Description: | PEO, myopathy, ataxia. | | Mutations: | L304R, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 45, Age of Patient: 74, Age of Death: n/a |
| Reference: | Lamantea et al, 2002; | | Description: | PEO. cytochrome c oxidase negative ragged red fibers, and multiple mtDNA deletions in skeletal muscle. | | Mutations: | G848S, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Lamantea et al, 2002; | | Description: | PEO. cytochrome c oxidase negative ragged red fibers, and multiple mtDNA deletions in skeletal muscle. | | Mutations: | P587L | | SNPs: | PNF, T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Lamantea et al, 2002; | | Description: | PEO. cytochrome c oxidase negative ragged red fibers, and multiple mtDNA deletions in skeletal muscle. | | Mutations: | P587L, R309L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Sarzi et al, 2007; | | Description: | Alpers, trunk hypotonia, seizures at 4 months, RC deficiency in liver. 28% mtDNA copy number in liver. | | Mutations: | C224Y, P587L | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.3, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Stewart et al, 2009; | | Description: | Mild bilateral ptosis, PEO. | | Mutations: | P587L, P587L | | SNPs: | T251I, T251I | | Age group: | adult | | Age of Onset: 41, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Stewart et al, 2009; | | Description: | PEO, ptosis, proximal weakness, multiple mtDNA deletions-LPCR. 25% COX-deficient fibers | | Mutations: | A467T, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 45, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Taanman et al, 2009; | | Description: | Vomiting, FTT, hypotonic, lactic acidemia, hypoglycemia, bilateral dense cataracts, hepatomegaly, jaundice, septicemia. infantile hepatocerebral mtDNA depletion. Mother with E1136K allele reported as asymptomatic. Father with T251I/P587L allele reported as asymptomatic. | | Mutations: | E1136K, P587L | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.01, Age of Patient: n/a, Age of Death: 0.5 |
| Reference: | Tang et al, 2011; | | Description: | N/A (as reported in Tang 2011 JMG) | | Mutations: | N1157S, P587L | | SNPs: | T251I | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 9, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Ocular bulbar weakness, peripheral neuropathy, muscle weakness, CPEO, ptosis, hypothyroidism, abnormal muscle histology, abnormal muscle ultrastructure, large mitochondrial proliferation, ragged red fibers. 96% mtDNA copy number in blood. Patient II-44. | | Mutations: | G848S, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 81, Age of Death: n/a |
Back to top | Reference: | Tang et al, 2011; | | Description: | Peripheral neuropathy, exercise intolerance, muscle cramps after exercise, easy fatigability, arrythmia, CPEO, abnormal EMG/NCV, ptosis, cataract, abnormal muscle histology, abnormal muscle ultrastructure, COX deficiency, ragged red fibers. 123% mtDNA copy number in blood. | | Mutations: | H932Y, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 31, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Peripheral neuropathy, ptosis, muscle weakness, CPK abnormalities. 77% mtDNA copy number in blood. | | Mutations: | H932Y, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 41, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Stroke/ischaemic episodes, peripheral neuropathy, CPEO, abnormal EMG/NCV, ptosis, muscle weakness, headache/migraine. 167% mtDNA copy number in blood. | | Mutations: | K1191N, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 39, Age of Death: n/a |
| Reference: | Van Goethem et al, 2003c; | | Description: | MNGIE-like, presented with GI symptoms, ptosis, neuropathy, muscle weakness. | | Mutations: | N864S, P587L | | SNPs: | T251I | | Age group: | juvenile | | Age of Onset: 15, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Alpers | | Mutations: | G848S, P587L | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Wong et al, 2008; | | Description: | Developmental delay, dementia, lactic acidosis, microcephaly, FTT, hearing loss, abnormal MRI. | | Mutations: | P587L, R853Q | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 0.2, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Developmental delay/dementia, liver dysfunction, lactic acidosis, fatigue, pancreatitis, cyclic vomiting. | | Mutations: | K1191R, P587L | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Kollberg et al, 2005; | | Description: | PEO, severe ophthalmoplegia and ptosis, which appeared insidiously and had been present clinically since the age of 55. Muscle biopsy at age 68 revealed mitochondrial myopathy, as well as muscle inflammation with granulomas. multiple mtDNA deletions in muscle tissue. | | Mutations: | G848S, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 55, Age of Patient: 75, Age of Death: n/a |
| Reference: | Burusnukul and de los Reyes, 2009; | | Description: | Global developmental delay, Central hypotonia, Torticollis, feeding difficulty at birth, myoclonic seizure, horizontal nystagmus, Intermittent estropia, Abnormal MRI | | Mutations: | P587L | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.4, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Lutz et al, 2009; | | Description: | Presented with feeding difficulties and hypotonia, At 4.5 months of age the patient contracted an influenza A infection. Concurrently she experienced hepatomegaly associated with severe hypoglycemia, Abnormalities in liver synthetic function, elevations in liver hepatocellular enzymes, elevated cerebrospinal fluid (CSF) protein, severely reduced mtDNA (3%) in liver. liver failure. She showed signs of ongoing respiratory difficulty, pancreatitis, and renal tubulopathy | | Mutations: | K1191R, P587L | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.166, Age of Patient: 0.416, Age of Death: 0.458 |
Back to top | Reference: | Weiss and Saneto, 2010; | | Description: | progressively droopy eyelids, double vision, gait instability, proximal arm weakness, ragged red and ragged blue (succinate dehydrogenase–positive) fibers in about 3% and 6% of all muscle fibers, bilateral ptosis, diplopia, dysarthria, myopathy involving the facial muscles, sensory ataxic polyneuropathy | | Mutations: | G848S, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 80, Age of Death: n/a |
| Reference: | Amiot et al, 2009; | | Description: | Peripheral neuropathy, PEO, ataxia, epilepsy, fatty liver | | Mutations: | D1184N, P587L | | SNPs: | T251I | | Age group: | childhood | | Age of Onset: 9, Age of Patient: n/a, Age of Death: 33 |
| Reference: | Amiot et al, 2009; | | Description: | Peripheral neuropathy, PEO, fatty liver | | Mutations: | D1184N, P587L | | SNPs: | T251I | | Age group: | childhood | | Age of Onset: 9, Age of Patient: n/a, Age of Death: 30 |
| Reference: | Aitken et al, 2009; | | Description: | Progressively blurred vision, diplopia, and longstanding bilateral ptosis (adPEO). She described occasional choking episodes after eating as well as fatigue and shortness of breath after minimal exertion. Lower limb examination revealed symmetric proximal limb weakness with reduced reflexes and flexor plantars. Tan- dem gait was hesitant. The initial differential diagnosis included Graves thyroid eye disease, a neuromuscular junction disor- der (myasthenia gravis or botulism), oculopharyngeal muscular dystrophy, Miller Fisher variant of Guillain-Barre syndrome, and progressive muscular dystrophy. Borderline myopathy. The muscle biopsy showed 13% cytochrome c ox- idase (COX)–deficient fibers, significant numbers of ragged red fibers but no excess of lipid or glycogen accumulation, and subsarcolemmal accumulation of abnormal mitochondria suggestive of a mitochondrial cytopathy. The patient presented 10 months later with an un- pleasant “jumping†sensation in her feet when at rest which was relieved by movement. Symptoms were worse at night and she also described sudden involuntary movements of her lower limbs (restless legs syndrome, RLS). | | Mutations: | P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 53, Age of Patient: 58, Age of Death: n/a |
| Reference: | Hanisch et al, 2014; | | Description: | Pareses, ptosis, ataxia, sensory neoropathy, motor neuropathy, axonal neuropathy, demyelinating neuropathy. | | Mutations: | P587L, R869Q | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 29, Age of Patient: 34, Age of Death: n/a |
Back to top | Reference: | Harris et al, 2010; | | Description: | Progressive ataxia, asymmetrical right ophthalmoplegia and encephalopathy. Seizures. Loss of extraocular movement on the right and right arm and leg spasticity and hyperreflexia. Elevated lactate level, a markedly depressed N-acetylaspartate level, and a markedly elevated choline level. Steroid-induced hyperglycemia. Progressive acute disseminated encephalomyelitis (ADEM). | | Mutations: | P587L | | SNPs: | T251I | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 4, Age of Death: 4.75 |
| Reference: | Lovan et al, 2013; | | Description: | diabetes mellitus 2, congestive heart failure, hypothyroidism, hypertension, depression with psychotic features and gastric bypass surgery. gait difficulties, cognitive impairment, ophthalmoplegia, resting tremor and peripheral neuropathy. Shooting pains and numbness in her lower extremities, occasional rectal incontinence. Presyncopal dizziness. Complicated diabetic retinopathy. sensory ataxia secondary to a sensorimotor polyneuropathy, chronic supranuclear ophthalmoplegia, and a resting tremor of her right hand. bilateral ptosis and marked temporal muscle wasting. Her sensory examination found loss of pinprick sensation, temperature and vibration bilaterally up to her hips. She had bilateral vibration loss at her fingertips. She had loss of proprioception at her toes. She had bilateral dysdiadochokinesia with significant ataxia on finger to nose testing. Arreflexic in her lower extremities. On cognitive testing she was found to have moderate dementia with significant deficits in registration and construction as well as ideomotor apraxia. | | Mutations: | A467T, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 55, Age of Patient: 60, Age of Death: n/a |
| Reference: | Scuderi et al, 2015; | | Description: | He presented migraine by age 7 and complained of fatigue during exercise by age 12. mild diffuse muscle hypotonia and flat feet. ptosis at 14. Brain MRI showed mild white matter hyperintensity. | | Mutations: | P587L | | SNPs: | P116Q, T251I | | Age group: | childhood | | Age of Onset: 7, Age of Patient: 14, Age of Death: n/a |
| Reference: | Scuderi et al, 2015; | | Description: | Presented deafness from childhood. migraine and fatigue. On neurological examination she had mild diffuse muscle weakness. Audiometric examination disclosed neurosensorial hypoacusia. mild anxiety with specific phobias concerning indoor environment and crowd. | | Mutations: | P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 29, Age of Patient: 37, Age of Death: n/a |
| Reference: | Scuderi et al, 2015; | | Description: | delayed psychomotor development. At age 3, neurological examination showed muscular hypotonia, joint laxity, absent deep tendon reflexes, broadbased gait, scapular winging, accentuation of lumbar lordosis and flat feet. intellectual disability. progressive cognitive impairment. mild myopathy. | | Mutations: | P587L | | SNPs: | P116Q, T251I | | Age group: | childhood | | Age of Onset: 3, Age of Patient: 8, Age of Death: n/a |
Back to top | Reference: | Scuderi et al, 2015; | | Description: | leg pain after physical activity. mild diffuse muscle hypotonia, hypotrophy, mild diffuse muscle weakness, joint laxity, scapular winging and increase of lumbar lordosis. Borderline intellectual functioning. | | Mutations: | P587L | | SNPs: | T251I | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 10, Age of Death: n/a |
| Reference: | Stewart et al, 2011; | | Description: | Alpers. nocturnal nausea followed by loss of consciousness without motor relinquishing. obsessive–compulsive disorder. | | Mutations: | P587L, P587L | | SNPs: | T251I, T251I | | Age group: | juvenile | | Age of Onset: 16, Age of Patient: 26, Age of Death: n/a |
| Reference: | Del Bo et al, 2003; | | Description: | PEO, complicated by dysphagia, myopathy. | | Mutations: | P587L | | SNPs: | PNF, T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 52, Age of Death: n/a |
| Reference: | Del Bo et al, 2003; | | Description: | PEO, complicated by dysphagia, myopathy. | | Mutations: | P587L, R807P | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 71, Age of Death: n/a |
| Reference: | Miguel et al, 2014; | | Description: | slowly progressive bilateral blepharoptosis, severe PEO, dysphagia, facial diparesis and exercise intolerance, presented, at the age of 60, with left-dominant postural tremor, reduced left arm swing during gait, postural instability, axonal sensory polyneuropathy, CT showed diffuse cerebral atrophy. Parkinsons. | | Mutations: | P587L | | SNPs: | PNF, T251I | | Age group: | adult | | Age of Onset: 46, Age of Patient: 66, Age of Death: n/a |
Back to top | Reference: | Ashley et al, 2008; | | Description: | Encephalopathy onset 16 years of age, presented with epilepsy. | | Mutations: | P587L, P589L, W748S | | Age group: | juvenile | | Age of Onset: 16, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Tzoulis et al, 2009; | | Description: | PEO, ptosis, muscle fatigue, diplopia, a mild external ophthalmoplegia affecting horizontal, but not vertical gaze. Tendonreflexes were diminished in the lower extremities and a distal sensory defect in stocking distribution was seen. | | Mutations: | P587L, W748S | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 45, Age of Patient: 50, Age of Death: n/a |
| Reference: | Ylönen et al, 2013; | | Description: | Akinetic-rigid Parkinsons. impaired balance, freezing, and increased salivation. Constipation and muscle cramps. He had arterial hypertension and mild normocytic anemia. cytochrome c oxidase-negative muscle fibers. | | Mutations: | P587L, W748S | | SNPs: | E1142G, T251I | | Age group: | adult | | Age of Onset: 49, Age of Patient: 65, Age of Death: n/a |
| Reference: | Horvath et al, 2006; | | Description: | PEO with myopathy | | Mutations: | P587L, V1106I | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 35, Age of Death: n/a |
|
| T251I | | | | Number of patients: (with T251I) | 53 | | Found together with: | PNF=Putatively Non-Functional enzyme |  | Show Patient Data |
| Patient data are sorted by mutation combination frequency. | Reference: | Blok et al, 2009; | | Description: | Exercise intolerance, CPEO, retinitis pigmentosa, diabetes, limb girdle weakness. 2nd patient with cataract and myopathy | | Mutations: | A467T, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 51, Age of Death: n/a |
| Reference: | Gonzalez-Vioque et al, 2006; | | Description: | PEO with ptosis, mild atrial hypertrophy. Sister had PEO. | | Mutations: | M603L, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 61, Age of Patient: 65, Age of Death: n/a |
| Reference: | Horvath et al, 2006; | | Description: | PEO, Neuropathy. Affect sibling. | | Mutations: | L304R, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 60, Age of Patient: 68, Age of Death: n/a |
| Reference: | Horvath et al, 2006; | | Description: | PEO, myopathy, ataxia. | | Mutations: | L304R, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 45, Age of Patient: 74, Age of Death: n/a |
Back to top | Reference: | Lamantea et al, 2002; | | Description: | PEO. cytochrome c oxidase negative ragged red fibers, and multiple mtDNA deletions in skeletal muscle. | | Mutations: | G848S, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Sarzi et al, 2007; | | Description: | Alpers, trunk hypotonia, seizures at 4 months, RC deficiency in liver. 28% mtDNA copy number in liver. | | Mutations: | C224Y, P587L | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.3, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Stewart et al, 2009; | | Description: | PEO, ptosis, proximal weakness, multiple mtDNA deletions-LPCR. 25% COX-deficient fibers | | Mutations: | A467T, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 45, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Taanman et al, 2009; | | Description: | Vomiting, FTT, hypotonic, lactic acidemia, hypoglycemia, bilateral dense cataracts, hepatomegaly, jaundice, septicemia. infantile hepatocerebral mtDNA depletion. Mother with E1136K allele reported as asymptomatic. Father with T251I/P587L allele reported as asymptomatic. | | Mutations: | E1136K, P587L | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.01, Age of Patient: n/a, Age of Death: 0.5 |
| Reference: | Tang et al, 2011; | | Description: | N/A (as reported in Tang 2011 JMG) | | Mutations: | N1157S, P587L | | SNPs: | T251I | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 9, Age of Death: n/a |
Back to top | Reference: | Tang et al, 2011; | | Description: | Ocular bulbar weakness, peripheral neuropathy, muscle weakness, CPEO, ptosis, hypothyroidism, abnormal muscle histology, abnormal muscle ultrastructure, large mitochondrial proliferation, ragged red fibers. 96% mtDNA copy number in blood. Patient II-44. | | Mutations: | G848S, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 81, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Peripheral neuropathy, exercise intolerance, muscle cramps after exercise, easy fatigability, arrythmia, CPEO, abnormal EMG/NCV, ptosis, cataract, abnormal muscle histology, abnormal muscle ultrastructure, COX deficiency, ragged red fibers. 123% mtDNA copy number in blood. | | Mutations: | H932Y, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 31, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Peripheral neuropathy, ptosis, muscle weakness, CPK abnormalities. 77% mtDNA copy number in blood. | | Mutations: | H932Y, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 41, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Stroke/ischaemic episodes, peripheral neuropathy, CPEO, abnormal EMG/NCV, ptosis, muscle weakness, headache/migraine. 167% mtDNA copy number in blood. | | Mutations: | K1191N, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 39, Age of Death: n/a |
| Reference: | Van Goethem et al, 2003c; | | Description: | MNGIE-like, presented with GI symptoms, ptosis, neuropathy, muscle weakness. | | Mutations: | N864S, P587L | | SNPs: | T251I | | Age group: | juvenile | | Age of Onset: 15, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Wong et al, 2008; | | Description: | Alpers | | Mutations: | G848S, P587L | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Developmental delay/dementia, liver dysfunction, lactic acidosis, fatigue, pancreatitis, cyclic vomiting. | | Mutations: | K1191R, P587L | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Kollberg et al, 2005; | | Description: | PEO, severe ophthalmoplegia and ptosis, which appeared insidiously and had been present clinically since the age of 55. Muscle biopsy at age 68 revealed mitochondrial myopathy, as well as muscle inflammation with granulomas. multiple mtDNA deletions in muscle tissue. | | Mutations: | G848S, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 55, Age of Patient: 75, Age of Death: n/a |
| Reference: | Burusnukul and de los Reyes, 2009; | | Description: | Global developmental delay, Central hypotonia, Torticollis, feeding difficulty at birth, myoclonic seizure, horizontal nystagmus, Intermittent estropia, Abnormal MRI | | Mutations: | P587L | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.4, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Lutz et al, 2009; | | Description: | Presented with feeding difficulties and hypotonia, At 4.5 months of age the patient contracted an influenza A infection. Concurrently she experienced hepatomegaly associated with severe hypoglycemia, Abnormalities in liver synthetic function, elevations in liver hepatocellular enzymes, elevated cerebrospinal fluid (CSF) protein, severely reduced mtDNA (3%) in liver. liver failure. She showed signs of ongoing respiratory difficulty, pancreatitis, and renal tubulopathy | | Mutations: | K1191R, P587L | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.166, Age of Patient: 0.416, Age of Death: 0.458 |
Back to top | Reference: | Weiss and Saneto, 2010; | | Description: | progressively droopy eyelids, double vision, gait instability, proximal arm weakness, ragged red and ragged blue (succinate dehydrogenase–positive) fibers in about 3% and 6% of all muscle fibers, bilateral ptosis, diplopia, dysarthria, myopathy involving the facial muscles, sensory ataxic polyneuropathy | | Mutations: | G848S, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 80, Age of Death: n/a |
| Reference: | Amiot et al, 2009; | | Description: | Peripheral neuropathy, PEO, ataxia, epilepsy, fatty liver | | Mutations: | D1184N, P587L | | SNPs: | T251I | | Age group: | childhood | | Age of Onset: 9, Age of Patient: n/a, Age of Death: 33 |
| Reference: | Amiot et al, 2009; | | Description: | Peripheral neuropathy, PEO, fatty liver | | Mutations: | D1184N, P587L | | SNPs: | T251I | | Age group: | childhood | | Age of Onset: 9, Age of Patient: n/a, Age of Death: 30 |
| Reference: | Aitken et al, 2009; | | Description: | Progressively blurred vision, diplopia, and longstanding bilateral ptosis (adPEO). She described occasional choking episodes after eating as well as fatigue and shortness of breath after minimal exertion. Lower limb examination revealed symmetric proximal limb weakness with reduced reflexes and flexor plantars. Tan- dem gait was hesitant. The initial differential diagnosis included Graves thyroid eye disease, a neuromuscular junction disor- der (myasthenia gravis or botulism), oculopharyngeal muscular dystrophy, Miller Fisher variant of Guillain-Barre syndrome, and progressive muscular dystrophy. Borderline myopathy. The muscle biopsy showed 13% cytochrome c ox- idase (COX)–deficient fibers, significant numbers of ragged red fibers but no excess of lipid or glycogen accumulation, and subsarcolemmal accumulation of abnormal mitochondria suggestive of a mitochondrial cytopathy. The patient presented 10 months later with an un- pleasant “jumping†sensation in her feet when at rest which was relieved by movement. Symptoms were worse at night and she also described sudden involuntary movements of her lower limbs (restless legs syndrome, RLS). | | Mutations: | P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 53, Age of Patient: 58, Age of Death: n/a |
| Reference: | Harris et al, 2010; | | Description: | Progressive ataxia, asymmetrical right ophthalmoplegia and encephalopathy. Seizures. Loss of extraocular movement on the right and right arm and leg spasticity and hyperreflexia. Elevated lactate level, a markedly depressed N-acetylaspartate level, and a markedly elevated choline level. Steroid-induced hyperglycemia. Progressive acute disseminated encephalomyelitis (ADEM). | | Mutations: | P587L | | SNPs: | T251I | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 4, Age of Death: 4.75 |
Back to top | Reference: | Lovan et al, 2013; | | Description: | diabetes mellitus 2, congestive heart failure, hypothyroidism, hypertension, depression with psychotic features and gastric bypass surgery. gait difficulties, cognitive impairment, ophthalmoplegia, resting tremor and peripheral neuropathy. Shooting pains and numbness in her lower extremities, occasional rectal incontinence. Presyncopal dizziness. Complicated diabetic retinopathy. sensory ataxia secondary to a sensorimotor polyneuropathy, chronic supranuclear ophthalmoplegia, and a resting tremor of her right hand. bilateral ptosis and marked temporal muscle wasting. Her sensory examination found loss of pinprick sensation, temperature and vibration bilaterally up to her hips. She had bilateral vibration loss at her fingertips. She had loss of proprioception at her toes. She had bilateral dysdiadochokinesia with significant ataxia on finger to nose testing. Arreflexic in her lower extremities. On cognitive testing she was found to have moderate dementia with significant deficits in registration and construction as well as ideomotor apraxia. | | Mutations: | A467T, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 55, Age of Patient: 60, Age of Death: n/a |
| Reference: | Scuderi et al, 2015; | | Description: | He presented migraine by age 7 and complained of fatigue during exercise by age 12. mild diffuse muscle hypotonia and flat feet. ptosis at 14. Brain MRI showed mild white matter hyperintensity. | | Mutations: | P587L | | SNPs: | P116Q, T251I | | Age group: | childhood | | Age of Onset: 7, Age of Patient: 14, Age of Death: n/a |
| Reference: | Scuderi et al, 2015; | | Description: | Presented deafness from childhood. migraine and fatigue. On neurological examination she had mild diffuse muscle weakness. Audiometric examination disclosed neurosensorial hypoacusia. mild anxiety with specific phobias concerning indoor environment and crowd. | | Mutations: | P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 29, Age of Patient: 37, Age of Death: n/a |
| Reference: | Scuderi et al, 2015; | | Description: | delayed psychomotor development. At age 3, neurological examination showed muscular hypotonia, joint laxity, absent deep tendon reflexes, broadbased gait, scapular winging, accentuation of lumbar lordosis and flat feet. intellectual disability. progressive cognitive impairment. mild myopathy. | | Mutations: | P587L | | SNPs: | P116Q, T251I | | Age group: | childhood | | Age of Onset: 3, Age of Patient: 8, Age of Death: n/a |
| Reference: | Scuderi et al, 2015; | | Description: | leg pain after physical activity. mild diffuse muscle hypotonia, hypotrophy, mild diffuse muscle weakness, joint laxity, scapular winging and increase of lumbar lordosis. Borderline intellectual functioning. | | Mutations: | P587L | | SNPs: | T251I | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 10, Age of Death: n/a |
Back to top | Reference: | Rouzier et al, 2013; | | Description: | CPEO, multiple mtDNa deletions. | | Mutations: | G848S | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 45, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Horvath et al, 2006; | | Description: | PEO with myopathy | | Mutations: | P587L, P587L | | SNPs: | T251I, T251I | | Age group: | adult | | Age of Onset: 62, Age of Patient: 70, Age of Death: n/a |
| Reference: | Horvath et al, 2006; | | Description: | PEO with myopathy | | Mutations: | P587L, P587L | | SNPs: | T251I, T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 56, Age of Death: n/a |
| Reference: | Horvath et al, 2006; | | Description: | PEO with myopathy, chronic bronchitis. | | Mutations: | P587L, P587L | | SNPs: | T251I, T251I | | Age group: | adult | | Age of Onset: 63, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Stewart et al, 2009; | | Description: | Mild bilateral ptosis, PEO. | | Mutations: | P587L, P587L | | SNPs: | T251I, T251I | | Age group: | adult | | Age of Onset: 41, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Stewart et al, 2011; | | Description: | Alpers. nocturnal nausea followed by loss of consciousness without motor relinquishing. obsessive–compulsive disorder. | | Mutations: | P587L, P587L | | SNPs: | T251I, T251I | | Age group: | juvenile | | Age of Onset: 16, Age of Patient: 26, Age of Death: n/a |
| Reference: | Di Fonzo et al, 2003; | | Description: | PEO, axonal sensorimotor polyneuropathy | | Mutations: | P587L, R807P | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 60, Age of Patient: 71, Age of Death: n/a |
| Reference: | Di Fonzo et al, 2003; | | Description: | PEO, dysphagia | | Mutations: | P587L, R807P | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 45, Age of Patient: 52, Age of Death: n/a |
| Reference: | Del Bo et al, 2003; | | Description: | PEO, complicated by dysphagia, myopathy. | | Mutations: | P587L, R807P | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 71, Age of Death: n/a |
Back to top | Reference: | Lamantea et al, 2002; | | Description: | PEO. cytochrome c oxidase negative ragged red fibers, and multiple mtDNA deletions in skeletal muscle. | | Mutations: | P587L | | SNPs: | PNF, T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Del Bo et al, 2003; | | Description: | PEO, complicated by dysphagia, myopathy. | | Mutations: | P587L | | SNPs: | PNF, T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 52, Age of Death: n/a |
| Reference: | Miguel et al, 2014; | | Description: | slowly progressive bilateral blepharoptosis, severe PEO, dysphagia, facial diparesis and exercise intolerance, presented, at the age of 60, with left-dominant postural tremor, reduced left arm swing during gait, postural instability, axonal sensory polyneuropathy, CT showed diffuse cerebral atrophy. Parkinsons. | | Mutations: | P587L | | SNPs: | PNF, T251I | | Age group: | adult | | Age of Onset: 46, Age of Patient: 66, Age of Death: n/a |
| Reference: | Agostino et al, 2003; | | Description: | PEO, bilateral ptosis, severe limita- tion of ocular motility, and a mosaic distribution of ragged-red and cytochrome c oxidase–negative fibers in the muscle biopsy. All patients had multiple deletions of muscle mtDNA. | | Mutations: | P587L, R227W | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 48, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Ashley et al, 2008; | | Description: | Alpers, epilepsy and hepatopathy, onset 5 months of age. | | Mutations: | P587L, R232G | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Ferrari et al, 2005; | | Description: | Infantile hepatocerebral, presented at 3 months as hypotonia followed by mtDNA depletion in liver, dementia, and death at 6 months by ventilatory insufficiency. | | Mutations: | P587L, R232G | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: 0.3, Age of Patient: n/a, Age of Death: 0.5 |
| Reference: | Tzoulis et al, 2009; | | Description: | PEO, ptosis, muscle fatigue, diplopia, a mild external ophthalmoplegia affecting horizontal, but not vertical gaze. Tendonreflexes were diminished in the lower extremities and a distal sensory defect in stocking distribution was seen. | | Mutations: | P587L, W748S | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 45, Age of Patient: 50, Age of Death: n/a |
| Reference: | Ylönen et al, 2013; | | Description: | Akinetic-rigid Parkinsons. impaired balance, freezing, and increased salivation. Constipation and muscle cramps. He had arterial hypertension and mild normocytic anemia. cytochrome c oxidase-negative muscle fibers. | | Mutations: | P587L, W748S | | SNPs: | E1142G, T251I | | Age group: | adult | | Age of Onset: 49, Age of Patient: 65, Age of Death: n/a |
| Reference: | Horvath et al, 2006; | | Description: | PEO with myopathy | | Mutations: | P587L, V1106I | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 35, Age of Death: n/a |
Back to top | Reference: | Ferreira et al, 2011; | | Description: | Ptosis, oculopharyngeal muscular dystrophy-like, pigmentary retinopathy. | | Mutations: | P587L, P648R | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 59, Age of Patient: 67, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Developmental delay, dementia, lactic acidosis, microcephaly, FTT, hearing loss, abnormal MRI. | | Mutations: | P587L, R853Q | | SNPs: | T251I | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 0.2, Age of Death: n/a |
| Reference: | Hanisch et al, 2014; | | Description: | Pareses, ptosis, ataxia, sensory neoropathy, motor neuropathy, axonal neuropathy, demyelinating neuropathy. | | Mutations: | P587L, R869Q | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 29, Age of Patient: 34, Age of Death: n/a |
| Reference: | Lamantea et al, 2002; | | Description: | PEO. cytochrome c oxidase negative ragged red fibers, and multiple mtDNA deletions in skeletal muscle. | | Mutations: | P587L, R309L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: n/a, Age of Death: n/a |
|
| A467T | | | | Number of patients: (with A467T) | 236 | | Found as the only mutation: | 3% of entries (6 patients) | | Found together with: | PNF=Putatively Non-Functional enzyme |  | Show Patient Data |
| Patient data are sorted by mutation combination frequency. | Reference: | Ashley et al, 2008; | | Description: | Epilepsy, ataxia | | Mutations: | A467T, A467T | | Age group: | juvenile | | Age of Onset: 16, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Stewart et al, 2009; | | Description: | Alpers, Encephalopathy, liver failure. | | Mutations: | A467T, A467T | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Stewart et al, 2009; | | Description: | PEO, ataxia, ataxic sensory axonal neuropathy, dysarthria, multiple mtDNA deletions. 10% COX deficient fibers, 2% RRF. | | Mutations: | A467T, A467T | | Age group: | adult | | Age of Onset: 30, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Stewart et al, 2009; | | Description: | PEO, ataxia, seizures, encephalopathy, ptosis, sensory neuropathy, mtDNA multiple deletions. 20% COX deficient fibers, 3% ragged red fibers. | | Mutations: | A467T, A467T | | Age group: | childhood | | Age of Onset: 12, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Tang et al, 2011; | | Description: | Age 19.6, developmental delay, after 18 years, rapid onset of muscle weakness, ataxia, myoclonic seizures, optic atrophy, diplopia, dysarthria. 102% mtDNA copy number in blood. | | Mutations: | A467T, A467T | | Age group: | juvenile | | Age of Onset: 18, Age of Patient: 20, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Ataxia, peripheral neuropathy, muscle weakness, easy fatigability, CPEO, abnormal EMG/NCV, ptosis, delayed gastric emptying, diarrhoea, constipation, lactic acidosis, abnormal muscle ultratstructure, ragged red fibers. 75% mtDNA copy number in blood. | | Mutations: | A467T, A467T | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 46, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Stroke/ischaemic episodes, ataxia, seizures, myoclonic seizures, peripheral neuropathy, CPEO. 114% mtDNA copy number in blood. | | Mutations: | A467T, A467T | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 40, Age of Death: n/a |
| Reference: | Winterthun et al, 2005; | | Description: | At age 17, she had an epileptic seizure preceded by tiredness and blurred vision. Visual blurring and poor memory persisted and 5 days later she developed status epilepticus with a focal start in the right arm. Examination showed horizontal nystagmus that did not settle, normal eye movements, and normal peripheral findings except that the deep tendon reflexes were recorded as weak. She returned 3 years later at age 20 with headache and unsteadiness. myoclonus and progressive ophthalmoplegia, ataxia, frequent myoclonic jerks. Cognitive dysfunction, axonal neuropathy. | | Mutations: | A467T, A467T | | Age group: | juvenile | | Age of Onset: 17, Age of Patient: 49, Age of Death: n/a |
| Reference: | Winterthun et al, 2005; | | Description: | Headache, tremor, sensory ataxia, dysarthria, nystagmus, ophthalmoplegia, cognitive dysfunction, axonal neuropathy, demyelinating neuropathy. She had headaches with preceding visual symptoms diagnosed as migraine that started at age 16. Episodic, involuntary jerking movements involving head and hands developed soon after. At age 18 she had two tonic clonic seizures preceded by poor concentration, confusion, and increased involuntary movements. Examination recorded titubation and myoclonus of the arms diminished reflexes in the legs, and reduced proprioception at the hallux. She has recurrent seizures and frequent headaches. At age 26 cerebellar and sensory ataxia, dysarthria, limita- tion of horizontal and vertical eye movements, and absent reflexes were recorded. COX negative fibers. At age 37 she was admitted with status epilepticus preceded by headache and visual symptoms in the right visual field. She remains ataxic but ambulant with assistance. | | Mutations: | A467T, A467T | | Age group: | juvenile | | Age of Onset: 16, Age of Patient: 39, Age of Death: n/a |
Back to top | Reference: | Winterthun et al, 2005; | | Description: | Headache, tremor. At age 20, he developed unsteadiness and dizziness. Examination showed upper limb tremor and myoclonus, and titubation. At age 31 limitation of horizontal eye movements was recorded; muscle biopsy was reported as showing no ragged red fibers. Subsequently, he developed limb and truncal ataxia, worsening ophthalmoplegia, pain in his extremities with glove and stocking sensory loss, particularly affecting proprioception, and sudden falls with altered consciousness that responded to anticonvulsant treatment. Sensory ataxia. axonal neuropathy, ophthalmoplegia, demyelination neuropathy, cognitive dysfunction, focal occipital epilepsy. | | Mutations: | A467T, A467T | | Age group: | childhood | | Age of Onset: 10, Age of Patient: 43, Age of Death: n/a |
| Reference: | Winterthun et al, 2005; | | Description: | This man developed epilepsy at age 5. At age 16 he developed unsteadiness and was euphoric. He had a right-sided divergent squint and difficulties on upgaze when the right eye would drift outward. Eye movements were otherwise full. There were horizontal nystagmus, absent tendon reflexes, and loss of proprioception distally in the legs. His gait was ataxic. Positive Romberg sign. At age 20 mild dysarthria was noted. Subsequently, he developed finger dysmetria, dysdiadochokinesis and myoclonus involving his arms, diarrhea, weight loss and cachexia, and ophthalmoplegia. Between the ages of 35 and 54 years he had infrequent seizures, but at age 55 he developed treatment-resistant status epilepticus and died. Cognitive dysfunction, axonal neuropathy, status epilepticus. | | Mutations: | A467T, A467T | | Age group: | childhood | | Age of Onset: 5, Age of Patient: n/a, Age of Death: 55 |
| Reference: | Wong et al, 2008; | | Description: | Onset 23 years with ataxia, neuropathy, hearing loss, seizures, and VPA liver failure. From "Saneto et al, 2010": Sensory neuropathy, ataxia, seizure onset at 15 years, simple partial seizure and epilepsia partialis continua myoclonus, VPA treatment caused liver failure after 3 months, progressive encephalopathy, Sensorineural hearing loss, dysmetria, intention tremor, hypotonia | | Mutations: | A467T, A467T | | Age group: | juvenile | | Age of Onset: 15, Age of Patient: 23, Age of Death: 23.5 |
| Reference: | Wong et al, 2008; | | Description: | Onset 32 years with neuropathy, myopathy, SANDO, PEO. | | Mutations: | A467T, A467T | | Age group: | adult | | Age of Onset: 32, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Nguyen et al, 2005; | | Description: | Refractory seizures, psychomotor regression, liver disease, presented with epilepsia partialis continua, Transient lactic acidemia | | Mutations: | A467T, A467T | | Age group: | childhood | | Age of Onset: 8.5, Age of Patient: n/a, Age of Death: 9 |
Back to top | Reference: | Milone et al, 2011; | | Description: | progressive bilateral ptosis, limited eye movements, lower extremities paresthesias, and unsteadiness, Multiple mtDNA deletions detected by PCR in muscle | | Mutations: | A467T, A467T | | Age group: | adult | | Age of Onset: 31, Age of Patient: 34, Age of Death: n/a |
| Reference: | Wolf et al, 2009; | | Description: | Developemental delay, status epilepticus onset, valproic acid therapy 2 weeks, epilepsia partialis continua, mtDNA depletion in liver | | Mutations: | A467T, A467T | | Age group: | infantile | | Age of Onset: 2.67, Age of Patient: 6, Age of Death: n/a |
| Reference: | Wolf et al, 2009; | | Description: | status epilepticus onset, valproic acid therapy 12 weeks, liver failure, | | Mutations: | A467T, A467T | | Age group: | childhood | | Age of Onset: 9.25, Age of Patient: 11, Age of Death: n/a |
| Reference: | McHugh et al, 2010; | | Description: | progressive difficulty walking, hand and foot numbness, ataxia, dysarthria, ptosis, sensory ataxia, with progressive ophthalmoplegia, dysarthria, nystagmus, dysphagia. Dysarthria became evident from age 55 years, and she developed ptosis at 62 years. | | Mutations: | A467T, A467T | | Age group: | adult | | Age of Onset: 42, Age of Patient: 62, Age of Death: n/a |
| Reference: | McHugh et al, 2010; | | Description: | progressive imbalance, hand and foot numbness, with impotence and dysarthria, progressive diplopia, bilateral ptosis with severe ophthalmoparesis in all directions and diplopia on lateral gaze. There was mild dysarthria and severe sensory ataxia, mild weakness in proximal and distal muscle groups, myopathy, multiple mtDNA deletions in muscle. | | Mutations: | A467T, A467T | | Age group: | adult | | Age of Onset: 41, Age of Patient: 46, Age of Death: n/a |
Back to top | Reference: | Brinjikji et al, 2011; | | Description: | Elevated CSF protein, occipital stroke, visual field deficit, peripheral neuropathy, migraine, epilepsy, ataxia, myoclonus, progressive gait ataxia, ophthalmoplegia, dysarthria, dysphagia, valproate induced encephalopathy and depression | | Mutations: | A467T, A467T | | Age group: | juvenile | | Age of Onset: 18, Age of Patient: 39, Age of Death: n/a |
| Reference: | Brinjikji et al, 2011; | | Description: | Hospitalized at age 7 for headache, fever, lethargy, and seizures, Elevated CSF protein, occipital stroke, visual field deficit, peripheral neuropathy, migraine, epilepsy, The patient began valproate therapy at age 11 and suffered hepatic encephalopathy which resolved after carnitine therapy. | | Mutations: | A467T, A467T | | Age group: | childhood | | Age of Onset: 7, Age of Patient: 28, Age of Death: n/a |
| Reference: | Hunter et al, 2011; | | Description: | Myoclonic Seizures, Focal motor seizures, Epilepsia partialis continua, Developmental Delay or Regression, Abnormal Liver Enzymes, Alpers | | Mutations: | A467T, A467T | | Age group: | childhood | | Age of Onset: 5, Age of Patient: n/a, Age of Death: 10.83 |
| Reference: | Visser et al, 2011; | | Description: | Migraine, seizures, dysphasia, magnesium infusion stopped the seizures | | Mutations: | A467T, A467T | | Age group: | juvenile | | Age of Onset: 17, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Visser et al, 2011; | | Description: | Seizures, magnesium infusion stopped the seizures | | Mutations: | A467T, A467T | | Age group: | juvenile | | Age of Onset: 19, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Tang et al, 2012; | | Description: | ptosis, PEO, muscle weakness, fatigability, peripheral neuropathy, ataxia, lactic acidosis and diarrhea alternating with constipation, ragged-red fibers, | | Mutations: | A467T, A467T | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 46, Age of Death: n/a |
| Reference: | Lax et al, 2012a; | | Description: | CPEO, Ptosis, Peripheral neuropathy, COX-deficient fibers, presence of mitochondrial dna deletions in muscle, Severe sensory and moderate motor neuronopathy, Distal and proximal neurogenic change, | | Mutations: | A467T, A467T | | Age group: | adult | | Age of Onset: 41, Age of Patient: 44, Age of Death: n/a |
| Reference: | Lax et al, 2012a; | | Description: | CPEO, Ptosis, Peripheral neuropathy, COX-deficient fibers, ragged red fibers, presence of mitochondrial dna deletions in muscle, Sensory and motor neuronopathy, Distal neurogenic change, proximal myopathy. | | Mutations: | A467T, A467T | | Age group: | juvenile | | Age of Onset: 18, Age of Patient: 36, Age of Death: n/a |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with migraine like headaches, ataxia, epilepsy, status epilepticus, nystagmus, myoclonus, neuropathy | | Mutations: | A467T, A467T | | Age group: | juvenile | | Age of Onset: 15, Age of Patient: 20, Age of Death: n/a |
| Reference: | Boes et al, 2009; | | Description: | Presented with status epilepticus, cerebellar ataxia and myoclonus, epilepsia partialis continua, Transient liver dysfunction with sodium valproate treatment at age 15, refractory focal motor status at age 17, | | Mutations: | A467T, A467T | | Age group: | childhood | | Age of Onset: 5, Age of Patient: n/a, Age of Death: 17 |
Back to top | Reference: | Tzoulis et al, 2013; | | Description: | Epilepsy, stroke-like episodes, ataxia, peripheral neuropathy, progressive external ophthalmoplegia (PEO). | | Mutations: | A467T, A467T | | Age group: | juvenile | | Age of Onset: 16, Age of Patient: n/a, Age of Death: 53 |
| Reference: | Tzoulis et al, 2013; | | Description: | Epilepsy, ataxia, peripheral neuropathy, progressive external ophthalmoplegia (PEO). | | Mutations: | A467T, A467T | | Age group: | juvenile | | Age of Onset: 15, Age of Patient: 42, Age of Death: n/a |
| Reference: | Hanisch et al, 2014; | | Description: | Ataxia, ptosis, pareses, sensory neuropathy, motor neuropathy, axonal neuropathy | | Mutations: | A467T, A467T | | Age group: | adult | | Age of Onset: 38, Age of Patient: 40, Age of Death: n/a |
| Reference: | Tzoulis et al, 2014; | | Description: | Epilepsy, stroke-like episode, Ataxia, Neuropathy, PEO | | Mutations: | A467T, A467T | | Age group: | childhood | | Age of Onset: 8, Age of Patient: n/a, Age of Death: 47 |
| Reference: | Tzoulis et al, 2014; | | Description: | Epilepsy, stroke-like episode, Ataxia, Neuropathy, PEO | | Mutations: | A467T, A467T | | Age group: | juvenile | | Age of Onset: 15, Age of Patient: n/a, Age of Death: 44 |
Back to top | Reference: | Tzoulis et al, 2014; | | Description: | Epilepsy, stroke-like episode, Ataxia, Neuropathy, PEO | | Mutations: | A467T, A467T | | Age group: | juvenile | | Age of Onset: 16, Age of Patient: n/a, Age of Death: 53 |
| Reference: | Schicks et al, 2010; | | Description: | early-onset ataxia, epilepsy, sensory neuropathy. | | Mutations: | A467T, A467T | | Age group: | childhood | | Age of Onset: 12.5, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Van Goethem et al, 2004; | | Description: | Increasing gait unsteadiness, mild cognitive decline in the fifth decade. Cataracts. ataxic gait. Deep tendon reflexes were absent. Vibration and position sense were absent at the lower limbs early on, whereas light touch sensation was decreased at a later stage. Pes Cavus. gaze paresis, nystagmus. axonal generalized peripheral neuropathy. brainstem dysfunction. | | Mutations: | A467T, A467T | | Age group: | juvenile | | Age of Onset: n/a, Age of Patient: 49, Age of Death: n/a |
| Reference: | Van Goethem et al, 2004; | | Description: | Increasing gait unsteadiness. ataxic gait. Deep tendon reflexes were absent. Vibration and position sense were absent at the lower limbs early on, whereas light touch sensation was decreased at a later stage. Pes Cavus. Dysarthria. horizontal and vertical gaze-evoked nystagmus. axonal generalized peripheral neuropathy. brainstem dysfunction. | | Mutations: | A467T, A467T | | Age group: | juvenile | | Age of Onset: n/a, Age of Patient: 47, Age of Death: n/a |
| Reference: | Van Goethem et al, 2004; | | Description: | at 18 year of age, he hadstatus epilepticus lasting 8 days, followed by a “Todd paralysis” of the left arm and face. Five years later, he had an acute psychiatric illness, hyperventilation, gastrointestinal symptoms, gait unsteadiness, and disturbed limb coordination. Between 32 and 35 years of age, he lost 13 kg of weight. On exami- nation, he had sensory gait ataxia, limb ataxia, areflexia, generalized dystrophy, and loss of vibration and static joint position sense at the distal lower limbs, severe dysarthria, and a left-sided Babinski sign. Romberg test was positive. intestinal pseudo-obstruction, anorexia, and further weight loss. A few weeks later, he developed stupor (Glasgow coma scale, 5/15), hyperventilation, myoclonic jerks, and seizures necessitating intensive care and artificial ventilation. gastroparesis. dilated cardiomyopathy. | | Mutations: | A467T, A467T | | Age group: | juvenile | | Age of Onset: 18, Age of Patient: n/a, Age of Death: 39 |
Back to top | Reference: | Rajakulendran et al, 2016; | | Description: | Alpers. Speech and motor delay were noted at the age of two years. At three years she developed epilepsy and six months later experienced migrainous attacks associated with vomiting, vertigo and transient left-sided weakness. At the age of five years she developed status epilepticus. A month later she was noted to have nystagmus, hypotonia of the lower limbs and absent knee jerks. Liver dysfunction. | | Mutations: | A467T, A467T | | Age group: | childhood | | Age of Onset: 3, Age of Patient: n/a, Age of Death: 5.5 |
| Reference: | Rajakulendran et al, 2016; | | Description: | MELAS. Occipital headaches. left homonymous hemianopia suggestive of a stroke-like episode. right occipital infarct. Jerking movements of her left arm suggestive of epilepsia partialis continua with dystonia, which was refractory to treatment. She developed an asymptomatic axonal neuropathy, deafness and myopathic weakness. bilateral ptosis, ophthalmoparesis, a dense left homonymous hemianopia, dysarthric speech, increased tone with clawing of the left hand, and distal muscle weakness. In addition, Romberg’s test was positive and she walked with a wide-based gait. ataxic gait. axonal sensory motor neuropathy. ragged red fibres and COX-negative fibres. | | Mutations: | A467T, A467T | | Age group: | adult | | Age of Onset: 24, Age of Patient: 31, Age of Death: n/a |
| Reference: | Rajakulendran et al, 2016; | | Description: | MEMSA. sensory neuropathy affecting legs. normal development until the age of 6 years when she presented with an encephalopathic illness consisting of impaired consciousness, vomiting and generalised tonic-clonic seizures. One week later she developed a left homonymous hemianopia. She remained stable until the age of 13 years when she developed stimulus sensitive myoclonus, tremor and a progressive cerebellar ataxia. Mildly elevated blood lactate and alanine levels and a sensory axonal peripheral neuropathy. occipital lobe infarcts. COX negative fibres. gaze-evoked nystagmus at 17. hand tremor, stimulus- sensitive myoclonus, head titubation, and gait ataxia. | | Mutations: | A467T, A467T | | Age group: | childhood | | Age of Onset: 6, Age of Patient: 16, Age of Death: n/a |
| Reference: | Rajakulendran et al, 2016; | | Description: | SANDO. Severe axonal neuropathy. COX-negative fibres, ragged red fibres. presented at the age of 20 years with diplopia and bilateral ptosis. Over the next five years he developed dysphagia, slurred speech and an unsteady gait. tingling sensation in hands, feet, leg, trunk and arms. at 44 years demonstrated bilateral ptosis and limitation of eye movements in all directions of gaze. dysarthria. Romberg’s test was positive. ataxic gait. axonal sensory peripheral neuropathy. ragged red fibres and more than 10 COX-negative fibres. | | Mutations: | A467T, A467T | | Age group: | adult | | Age of Onset: 20, Age of Patient: 44, Age of Death: n/a |
| Reference: | Janssen et al, 2016; | | Description: | At 21 years, presented with a first generalized tonic–clonic seizure. She complained of migraine afterward and developed a convulsive status epilepticus. Visual hallucinations, jerking of the right arm with secondary generalization. She died 7 months after initial presentation. | | Mutations: | A467T, A467T | | Age group: | adult | | Age of Onset: 21, Age of Patient: 21, Age of Death: 21.6 |
Back to top | Reference: | Janssen et al, 2016; | | Description: | Sudden falls, frequent negative myoclonus, ataxic gait and loss of proprioception in the distal extremities with nerve conduction studies demonstrating a mild axonal sensory polyneuropathy. myoclonic status epilepticus, generalized tonic–clonic seizure. | | Mutations: | A467T, A467T | | Age group: | childhood | | Age of Onset: 12, Age of Patient: 18, Age of Death: 27 |
| Reference: | Simonati et al, 2003a; | | Description: | During his early childhood, he had frequent “acetonaemic vomiting” and stunted growth. At age 7 years, he developed EPC and sensory ataxia. epilepsia partialis continua, followed by progressive ataxia Reduced density of the white matter. behavioural problems. Elevated blood lactate. myoclonic seizures. sensory ataxia, absent deep tendon reflexes, cerebellar dysfunction, nystagmus, peripheral vision defect, and a pale optic disk. mild atrophy of the frontal,parietal and visual cortices, focal hyperlucencies of the frontal and occipital cortex, as well as of the thalamus bilaterally, and cerebellar atrophy. COX-negative fibres. partial and generalized seizures as well as myoclonic fits. | | Mutations: | A467T, A467T | | Age group: | childhood | | Age of Onset: 7, Age of Patient: 18, Age of Death: 19 |
| Reference: | Blok et al, 2009; | | Description: | MELAS-like, features including occipital lobe epilepsy. | | Mutations: | A467T, W748S | | Age group: | juvenile | | Age of Onset: n/a, Age of Patient: 19, Age of Death: n/a |
| Reference: | Blok et al, 2009; | | Description: | Occipital lobe epilepsy, myoclonus, cognitive delay, polyneuropathy, cerebellar ataxia. | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 40, Age of Death: n/a |
| Reference: | Blok et al, 2009; | | Description: | Ptosis, CPEO, polyneuropathy, cerebellar ataxia, dysarthria. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 47, Age of Death: n/a |
Back to top | Reference: | Horvath et al, 2006; | | Description: | Onset at 34 years with PEO, ataxia, dysphagia, myopathy, neuropathy and Diabetes. | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 34, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Kollberg et al, 2006; | | Description: | Started as an ataxic syndrome, Alpers, ragged red fibers, COX-deficient fibers, mtDNA deletions. mtDNA copy numbers 1.6 fold. She was healthy and developed normally until her late preschool years when she developed slowly progressive ataxia. Occasional myoclonic seizures, and at 13 years of age, valproate treatment was started because of occipital seizures. ataxia, dementia, and cortical blindness. She developed refractory seizures, including multifocal EPC, status epilepticus, and myoclonus. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | childhood | | Age of Onset: 5, Age of Patient: 13, Age of Death: 14 |
| Reference: | Kollberg et al, 2006; | | Description: | psychomotor regression, refractory seizures, stroke-like episodes, hepatopathy, and ataxia. COX-deficient muscle fibers, mtDNA deletions, 160% mtDNA copy number. He was normal until 11 years of age when he developed mild tremor and ataxia. At 13 years of age, he reported occasional migraine-like headaches and was found to have mild ataxia and myoclonus. generalized seizures. mild leftsided hemiparesis, external ophthalmoplegia, and peripheral neuropathy. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | childhood | | Age of Onset: 11, Age of Patient: 24, Age of Death: n/a |
| Reference: | Sarzi et al, 2007; | | Description: | Feeding difficulties 2 days after birth and trunk hypotonia, dysmorphy, hypotonia, coloboma, heart failure at 2 months, liver insufficiency at 4 months. Liver and muscle mtDNA depletion. | | Mutations: | A467T, K561M, W748S | | Age group: | infantile | | Age of Onset: 0.001, Age of Patient: n/a, Age of Death: 0.5 |
| Reference: | Schulte et al, 2009; | | Description: | Dysarthria, sensory neuropathy, PEO, minor cognitive dysfunction, depression. | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 34, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Stewart et al, 2009; | | Description: | PEO, ataxia, seizures, ptosis, bilateral deafness, myopathy, dysarthria, peripheral neuropathy, multiple mtDNA deletions. 5% COX deficient fibers, 2% RRF. | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 39, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Autistic features, headaches/migraines, ataxia, seizures, intractable seizure, optic atrophy, abnormal MRI, dystonia, posterior stroke, abnormal EEG. 101% mtDNA copy number in blood. | | Mutations: | A467T, W748S | | Age group: | juvenile | | Age of Onset: n/a, Age of Patient: 18, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Hearing loss, Ataxia Neuropathy Spectrum, Seizures, dementia, developmental delay | | Mutations: | A467T, Q497H, W748S | | SNPs: | E1143G | | Age group: | juvenile | | Age of Onset: 17, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Hansen et al, 2012; | | Description: | gait instability, manual coordination disorder, speech disturbance and secondary generalized motor seizures, tension-type headaches, complete external ophthalmoplegia, cerebellar dysarthria and ataxia, sensory ataxia in Romberg-testing, areflexia of the lower extremities, mild cognitive dysfunction, epileptic seizures, generalised cortical atrophy, sensory axonal neuropathy, dysarthria | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 23, Age of Patient: 48, Age of Death: n/a |
| Reference: | Kinghorn et al, 2012; | | Description: | Ataxia, dysarthria, suffered two generalized seizures, and developed severe progressive cognitive decline and psychiatric manifestations including visual and auditory hallucinations, bilateral external ophthalmoplegia, bilateral ptosis, and reduced visual acuities, bilateral sensorineural hearing loss, proximal muscle weakness in all four limbs and was areflexic, severe sensory neuropathy and myopathy, | | Mutations: | A467T, W748S | | Age group: | juvenile | | Age of Onset: n/a, Age of Patient: 18, Age of Death: 65 |
Back to top | Reference: | Kinghorn et al, 2012; | | Description: | Diplopia, partial ophthalmoparesis in all directions of gaze and dysarthria, sensory neuropathy, COX-deficient fibers, multiple mtDNA deletions, | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 50, Age of Death: n/a |
| Reference: | Pelayo-Negro et al, 2012; | | Description: | ataxia, sensory neuropathy, dysarthria, and external ophthalmoparesis | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 30, Age of Patient: 50, Age of Death: n/a |
| Reference: | Lax et al, 2012a; | | Description: | CPEO, Ptosis, Peripheral neuropathy, COX-deficient fibers, presence of mitochondrial dna deletions in muscle, ataxia, Severe sensory, moderate motor axonal neuronopathy, Distal neurogenic, proximal myopathy | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 41, Age of Patient: 48, Age of Death: n/a |
| Reference: | Lax et al, 2012a; | | Description: | CPEO, Ptosis, Peripheral neuropathy, COX-deficient fibers, ragged red fibers, presence of mitochondrial dna deletions in muscle, Severe sensory and moderate motor neuronopathy, Distal neurogenic change, proximal myopathy | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 34, Age of Patient: 47, Age of Death: n/a |
| Reference: | Lax et al, 2012a; | | Description: | Peripheral neuropathy, ataxia, epilepsy, COX-deficient fibers, presence of mitochondrial dna deletions in muscle, Moderate sensory neuronopathy | | Mutations: | A467T, W748S | | Age group: | juvenile | | Age of Onset: 16, Age of Patient: 18, Age of Death: n/a |
Back to top | Reference: | Lax et al, 2012a; | | Description: | Peripheral neuropathy, ataxia, epilepsy, presence of mitochondrial dna deletions in muscle, | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 20, Age of Patient: 24, Age of Death: n/a |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with epilepsy and headaches, ataxia, status epilepticus, nystagmus, myoclonus, neuropathy. Sodium valproate treatment 6 weeks prior to death. Liver dysfunction, cause of death status epilepticus and liver failure. Hepatic histology showed acute liver necrosis. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | juvenile | | Age of Onset: 19, Age of Patient: n/a, Age of Death: 19 |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with epilepsy and headaches, ataxia, epilepsy, status epilepticus, myoclonus, neuropathy. Sodium valproate treatment 7 weeks prior to death. Liver dysfunction and rising liver enzymes and bilirubin terminally, cause of death status epilepticus. Hepatic histology showed centrolobular degeneration. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | juvenile | | Age of Onset: 14, Age of Patient: n/a, Age of Death: 23 |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with epilepsy, ataxia, status epilepticus, headaches, myoclonus, neuropathy. Sodium valproate treatment for 8 months. Stopped just prior to death. Liver dysfunction, cause of death liver failure. Hepatic histology showed acute liver necrosis. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | childhood | | Age of Onset: 10, Age of Patient: n/a, Age of Death: 10 |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with epilepsy, headaches, epilepsy, status epilepticus, headaches, nystagmus, and neuropathy. Sodium valproate treatment for 2 months then stopped 2 months prior to death. Liver dysfunction, liver transplant 1 month prior to death, cause of death liver failure. Hepatic histology showed acute liver necrosis. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: 20, Age of Patient: n/a, Age of Death: 20 |
Back to top | Reference: | Tzoulis et al, 2006; | | Description: | Presented with epilepsy, status epilepticus, headaches, nystagmus, myoclonus, neuropathy. Sodium valproate treatment 7 weeks prior to death. Liver dysfunction and rising liver enzymes and bilirubin terminally, cause of death status epilepticus. Hepatic histology showed centrolobular degeneration. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | juvenile | | Age of Onset: 15, Age of Patient: n/a, Age of Death: 21 |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with progressive gait unsteadiness, ataxia, headaches, myoclonus, neuropathy, PEO, ptosis onset at age 44 | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: 36, Age of Patient: 50, Age of Death: n/a |
| Reference: | Tzoulis et al, 2006; | | Description: | Presented with progressive gait unsteadiness, ataxia, headaches, neuropathy, PEO ptosis onset age 25 | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: 24, Age of Patient: 43, Age of Death: n/a |
| Reference: | Paus et al, 2008; | | Description: | Presented with PEO, mild bilateral ptosis, dysarthria, ataxia, and neuropathy | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: 36, Age of Patient: 37, Age of Death: n/a |
| Reference: | Paus et al, 2008; | | Description: | episodic headache, cognitive decline, and seizures, repeatedly resulting in generalized status epilepticus, ataxia, sensorimotor peripheral neuropathy, dysarthria, PEO with diplopia, mild bilateral ptosis, epilepsy, cognitive impairment, myoclonus, | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 28, Age of Patient: 39, Age of Death: n/a |
Back to top | Reference: | Rouzier et al, 2013; | | Description: | R-EPC (refractory epilepsia partialis continua), axonal neuropathy, cerebellar ataxia, hyperintensity of rolandic, occipital and cerebellar cortex and dentate nucleus. | | Mutations: | A467T, W748S | | Age group: | juvenile | | Age of Onset: 17, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Posada et al, 2010; | | Description: | Ataxic sensory neuropathy, dysarthria, progressive ophthalmoplegia, paresthesia of lower extremities, unsteady gait, slurred speech, absent deep tendon reflexes, vibrations, ataxic gait and positive Romberg sign. Multiple mtDNA deletions, a marked decrease in mtDNA copy number. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | adult | | Age of Onset: 37, Age of Patient: 47, Age of Death: n/a |
| Reference: | Roshal et al, 2011; | | Description: | Focal parieto-occipital lobe seizures, migraine headaches, cerebellar ataxia, sensory–motor axonal neuropathy, and impairment of visual perception and cognitive function. | | Mutations: | A467T, W748S | | Age group: | juvenile | | Age of Onset: 15, Age of Patient: 16, Age of Death: n/a |
| Reference: | Tuladhar et al, 2013; | | Description: | abnormal gait, ataxia, PEO, and jerky torticollis, mild cerebellar atrophy, sensory neuronopathy. | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 43, Age of Patient: 45, Age of Death: n/a |
| Reference: | Tzoulis et al, 2013; | | Description: | Ataxia, peripheral neuropathy, progressive external ophthalmoplegia (PEO). | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 24, Age of Patient: 50, Age of Death: n/a |
Back to top | Reference: | Tzoulis et al, 2013; | | Description: | Ataxia, peripheral neuropathy, progressive external ophthalmoplegia (PEO). | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 36, Age of Patient: 58, Age of Death: n/a |
| Reference: | Hanisch et al, 2014; | | Description: | Ataxia, ptosis, mild cognitive impairment, sensory neuropathy, motor neuropathy, axonal neuropathy, demyelinating neuropathy. | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 41, Age of Patient: 45, Age of Death: n/a |
| Reference: | Hanisch et al, 2014; | | Description: | Ataxia, ptosis, pareses, diabetes, sensory neuropathy, motor neuropathy, axonal neuropathy, demyelinating neuropathy. | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 49, Age of Patient: 56, Age of Death: n/a |
| Reference: | Lax et al, 2012b; | | Description: | Ataxia, Epilepsy, Dementia, Encephalopathy, Myoclonus, Fatigue, Depression, Dysarthria, Cardiac failure, cardiomyopathy, Diabetes, arPEO, Ischemic heart disease. | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 20, Age of Patient: 24, Age of Death: n/a |
| Reference: | Sitarz et al, 2014; | | Description: | Ataxia, neuropathy, PEO, MIRAS. | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 39, Age of Death: n/a |
Back to top | Reference: | Tzoulis et al, 2014; | | Description: | Epilepsy, stroke-like episode, Ataxia, Neuropathy. | | Mutations: | A467T, W748S | | Age group: | juvenile | | Age of Onset: 13, Age of Patient: n/a, Age of Death: 21 |
| Reference: | Tzoulis et al, 2014; | | Description: | Epilepsy, stroke-like episode, Ataxia, Neuropathy. | | Mutations: | A467T, W748S | | Age group: | juvenile | | Age of Onset: 14, Age of Patient: n/a, Age of Death: 23 |
| Reference: | Van Goethem et al, 2004; | | Description: | minor tremor of the hands from age 12 years. In his late 20s, he experienced tingling in both hands, followed by tingling in the legs. In the early fourth decade, he became unsteady at walking, especially in the dark. From his early 40s, his speech became slurred (dysarthria). On examination at age 52 years, he had an ataxic gait, and Romberg test was positive. There was dysarthria and bilateral ophthalmoparesis. generalized areflexia. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | childhood | | Age of Onset: 12, Age of Patient: 52, Age of Death: n/a |
| Reference: | Janssen et al, 2016; | | Description: | Migraine, ataxia, polyneuropathy. focal seizures with visual symptoms and motor signs, and secondary generalized tonic-clonic seizure. Clinical neurological examination revealed an ataxic gait and areflexia with sensory loss. Convulsive status epilepticus. Cerebral MRI revealed a left occipital and left thalamic lesion. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | juvenile | | Age of Onset: 17, Age of Patient: 20, Age of Death: 23 |
| Reference: | Janssen et al, 2016; | | Description: | Presented with status epilepticus, occipital lobe seizures. Further neurologic examination showed ataxic gait and appendicular ataxia. chronic sensory axonal polyneuropathy. Cognitive decline. At follow-up consultation 6 months after the initial presentation, she had not experienced any more seizures. | | Mutations: | A467T, W748S | | SNPs: | E1143G | | Age group: | juvenile | | Age of Onset: 18, Age of Patient: 18.7, Age of Death: n/a |
Back to top | Reference: | Janssen et al, 2016; | | Description: | She had occipital lobe and secondarily generalized seizures. In addition, she had PEO, truncal and appendicular ataxia and peripheral neuropathy with diminished vibration sense and areflexia. She reported sporadic occurrence of seizures with a frequency of once in every few months. | | Mutations: | A467T, W748S | | Age group: | adult | | Age of Onset: 21, Age of Patient: 47, Age of Death: n/a |
| Reference: | Ashley et al, 2008; | | Description: | Hepatopathy, Epilepsy, Hepatopathy, | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 1.5, Age of Patient: n/a, Age of Death: n/a |
| Reference: | de Vries et al, 2007; | | Description: | Onset at 6 months with FTT, dementia, hypotonia, seizures, hepatopathy, aplasia cutis, delayed myelinisation. Death at 15 months. | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: n/a, Age of Death: 1.3 |
| Reference: | Ferrari et al, 2005; | | Description: | He developed normally in the first 6 months. Refusal of food and failure to thrive were noted from the 7th month of life. At 1 year of age, increased transaminase levels in the blood were accompanied by liver enlargement and hyperecogenicity. Motor development was delayed. hypoglycaemic episodes and lactic acidosis. A liver biopsy at the age of 16 months revealed steatosis and fibrosis. At 20 months of age, he had numerous episodes of status epilepticus and epilepsia partialis continua. The patient died at 2.5 years of age during an episode of status epilepticus. Alpers’ syndrome. | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: n/a, Age of Death: 2.5 |
| Reference: | Kollberg et al, 2006; | | Description: | Ragged Red Fibers, COX-Deficient Fibers. The boy had feeding difficulties with failure to thrive since 3 months of age. Delayed motor development. general muscle weakness and ataxia. compromised liver function. | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 0.33, Age of Patient: n/a, Age of Death: 1.33 |
Back to top | Reference: | Stewart et al, 2009; | | Description: | Alpers, Epilepsy, developmental delay, COX deficient fibres. | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Stewart et al, 2009; | | Description: | Alpers, seizures, developmental delay, liver failure, hypotonia, impaired vision, renal stones, mtDNA depletion in muscle. 5% COX deficient fibers. | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 0.7, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Developmental delay, dementia, seizures, Alpers | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 2, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Developmental delay, dementia, seizures, Alpers, Ataxia | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 2, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Developmental delay, dementia, seizures, Alpers, Lactic acidosis, Cortical athrophy, hypoglycemia | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 1.5, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Wong et al, 2008; | | Description: | Developmental delay, dementia, seizures, Alpers, Lactic acidosis, Stroke | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 2, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Developmental delay, dementia, seizures, Alpers, Stroke, ataxia, exercise intolerance | | Mutations: | A467T, G848S | | Age group: | childhood | | Age of Onset: 9, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Developmental delay, dementia, seizures, Alpers, hypotonia, failure to thrive | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 0.8, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Nguyen et al, 2005; | | Description: | Refractory seizures, psychomotor regression, liver disease, Transient hypoglycemia, Transient lactic academia, Elevated CSF protein, presented with epilepsia partialis continua, | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 0.9, Age of Patient: n/a, Age of Death: 1.8 |
| Reference: | Nguyen et al, 2005; | | Description: | Refractory seizures, psychomotor regression, liver disease, presented with liver failure | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: 1.3 |
Back to top | Reference: | Saneto et al, 2010; | | Description: | Cyclic vomiting, episodic hypoglycemia then seizure onset at 6 years, generalized seizures, GI dysmotility, eyelid ptosis,ophthalmoplegia, sensorineural hearning loss, absent smooth pursuit eye movements | | Mutations: | A467T, G848S | | Age group: | childhood | | Age of Onset: 6, Age of Patient: 11, Age of Death: n/a |
| Reference: | Hunter et al, 2011; | | Description: | Myoclonic Seizures, Epilepsia partialis continua, Developmental Delay or Regression, ataxia, Abnormal Liver Enzymes, Serum Lactate, liver mtDNA depletion, Alpers | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: n/a, Age of Death: 3.33 |
| Reference: | Hunter et al, 2011; | | Description: | Myoclonic Seizures, Generalized tonic-clonic seizures, Focal motor seizures, Developmental Delay or Regression, hypotonia, tremor, Abnormal Liver Enzymes, Serum Lactate, liver mtDNA depletion, death by liver failure, Alpers | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 0.83, Age of Patient: n/a, Age of Death: 0.92 |
| Reference: | McCoy et al, 2011; | | Description: | focal seizures, elevated liver transaminases, Elevated CSF protein, patient died several weeks later | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 0.8, Age of Patient: n/a, Age of Death: 1 |
| Reference: | Scalais et al, 2012; | | Description: | presented with hypoglycemia, lactic acidosis, moderate ketosis and liver dysfunction, generalized hypotonia, progressive jaundice and abdominal distension with ascites, status epilepticus with generalized tonico-clonic seizures. She had an intermittent tremor, moderate truncal ataxia with a wide-based gait, myoclonic seizures, epilepsia partialis continua, | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 0.29, Age of Patient: n/a, Age of Death: 5 |
Back to top | Reference: | de Vries et al, 2008; | | Description: | Delayed motor and mental development, From the age of 3 years his psychomotor development declined and epilepsy, visual impairment and sensorineuronal deafness, epilepsia partialis continua. Bilateral ptosis, generalized hypotonia, ataxia, absent reflexes and hyperlaxity by age 5. | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 3, Age of Patient: 5, Age of Death: n/a |
| Reference: | Hasselmann et al, 2010; | | Description: | Alpers, status epilepticus and somnolence, ataxia with dysmetria and muscular hypotonia. Healthy unrelated parents. Laboratory investigations revealed myoclonic epilepsy with ragged red fibers, | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 2.2, Age of Patient: 3.5, Age of Death: 5.5 |
| Reference: | Tzoulis et al, 2013; | | Description: | Epilepsy, stroke-like episodes. | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 0.6, Age of Patient: n/a, Age of Death: 0.6 |
| Reference: | Roels et al, 2009; | | Description: | mtDNA depletion. severe hypoglycemia, liver biopsy shows micronodular cirrhosis. At 3 y 9 m, status epilepticus develops which occurs again at 4 y and is difficult to control. myoclonia. She dies from bronchopulmonary infection when 5 y old. COX negative fibers. | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 0.25, Age of Patient: 4, Age of Death: 5 |
| Reference: | Tzoulis et al, 2014; | | Description: | Epilepsy, stroke-like episode. | | Mutations: | A467T, G848S | | Age group: | infantile | | Age of Onset: 0.6, Age of Patient: n/a, Age of Death: 0.6 |
Back to top | Reference: | Ashley et al, 2008; | | Description: | Epilepsy, hepatopathy, movement disorder (present but not prominent) | | Mutations: | A467T | | SNPs: | PNF | | Age group: | infantile | | Age of Onset: 0.33, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Ashley et al, 2008; | | Description: | Epilepsy, hepatopathy, movement disorder (present but not prominent) | | Mutations: | A467T | | SNPs: | PNF | | Age group: | infantile | | Age of Onset: 0.58, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Ashley et al, 2008; | | Description: | mtDNA depletion (30% in muscle), epilepsy, hepatopathy, movement disorder | | Mutations: | A467T, PNF | | Age group: | infantile | | Age of Onset: 1.5, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Sarzi et al, 2007; | | Description: | Myoclonus epilepsy, Epilepsy, Alpers syndrome, steatosis, death at 3 years | | Mutations: | A467T | | SNPs: | PNF | | Age group: | childhood | | Age of Onset: 3, Age of Patient: n/a, Age of Death: 3 |
| Reference: | Sarzi et al, 2007; | | Description: | Seizures, Alpers syndrome, Jaundice at 9 months and liver enlargement. | | Mutations: | A467T | | SNPs: | PNF | | Age group: | infantile | | Age of Onset: 0.66, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Wong et al, 2008; | | Description: | Alpers, stroke, hypotonia, Dementia, developmental delay, seizures. | | Mutations: | A467T | | SNPs: | PNF | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Developmental delay, dementia, seizures, Alpers | | Mutations: | A467T | | SNPs: | PNF | | Age group: | childhood | | Age of Onset: 4, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Developmental delay, dementia, seizures, Alpers, Family history of acute liver failure | | Mutations: | A467T | | SNPs: | PNF | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Developmental delay, dementia, seizures, Alpers, Ptosis, ataxia, visual hallucinations | | Mutations: | A467T | | SNPs: | PNF | | Age group: | childhood | | Age of Onset: 3, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Nguyen et al, 2005; | | Description: | Refractory seizures, psychomotor regression, liver disease, presenting symptom was status epilepticus, Transient hypoglycemia, Transient lactic academia, hypercoagulable state | | Mutations: | A467T | | SNPs: | PNF | | Age group: | infantile | | Age of Onset: 0.8, Age of Patient: n/a, Age of Death: 1 |
Back to top | Reference: | Lutz et al, 2009; | | Description: | speech and motor delay at 1.5 yr, seizure that included eye deviation, jaw clenching, and hypotonia of the trunk and extremities. He experienced repetitive generalized tonic-clonic seizures that evolved into refractory status epilepticus, severe encephalopathy characterized by choreo-athetoid movements, corticovisual impairment, diffuse hypotonia, and severe swallowing dysfunction, elevated plasma lactate, CSF lactate and protein were both elevated, Liver transaminases remained mildly elevated. dysphagia. | | Mutations: | A467T | | SNPs: | PNF | | Age group: | infantile | | Age of Onset: 1.5, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Mousson de Camaret et al, 2011 | | Description: | Alpers. At 1 year of age, she presented a rightsided clonic status epilepticus resistant to multiple antiepileptic drugs. Carbamazepine worsened myoclonic jerks. A ketogenic diet was well tolerated but was ineffective. At 12 months, dramatic psychomotor regression was observed with poor interaction, major global hypotonia and no eye contact due to cortical blindness. epilepsia partialis continua reinforced by recurrent episodes of tonic clonic seizures. At 16 months of age, liver enlargement was noticed associated with mild cytolysis. The patient died at 27 months of age of respiratory failure. | | Mutations: | A467T | | SNPs: | PNF | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: n/a, Age of Death: 2.25 |
| Reference: | Naviaux and Nguyen, 2004; | | Description: | At 11 months, she developed otitis media and shortly thereafter she was admitted to the intensive care unit and intubated and treated for 2 days for apnea associated with an episode of absence status epilepticus. Brain computed tomography at that time was normal. Her gait was ataxic after recovery, and she had bouts of epilepsia partialis continua, initially involving the left hand. At 16 months, she suffered another episode of status epilepticus requiring intubation for airway protection and ventilatory support. Head control was lost permanently after this episode. she was found to have cortical blindness, and brain magnetic resonance imaging showed marked cerebral atrophy without focal abnormalities. A liver biopsy showed chronic and acute liver disease with micronodular cirrhosis, hepatocyte dropout, microvesicular fat, and bile ductular proliferation. The diagnosis of Alpers’ syndrome was made at that time. refractory partial seizures, scoliosis, quadriparesis, tracheomalacia, and osteoporosis and was aphasic. | | Mutations: | A467T, PNF, PNF | | Age group: | infantile | | Age of Onset: 0.9, Age of Patient: n/a, Age of Death: 10 |
| Reference: | Naviaux and Nguyen, 2004; | | Description: | He had been well and developmentally normal until 15 months when he suffered 3 days of a diarrheal illness and required admission for new ataxia and encephalopathy. On the day of admission, he developed status epilepticus and was intubated for 3 weeks for aspiration pneumonia. He developed progressive hepatic failure and cortical blindness. He remained in a vegetative state until he died. Autopsy showed massive cerebellar and cerebral atrophy. | | Mutations: | A467T, PNF, PNF | | Age group: | infantile | | Age of Onset: 1.25, Age of Patient: n/a, Age of Death: 1.3 |
| Reference: | Naviaux and Nguyen, 2004; | | Description: | His speech and motor development were normal until 19 months of age when he developed an episode of encephalopathy, ataxia, and hypertonia associated with an acute febrile illness. At 30 months, he developed a refractory focal myoclonic seizure disorder. His terminal admission was associated with a documented Rotavirus infection. At this time, he was noted to have cortical blindness, epilepsia partialis continua, and acute liver failure (hepatic failure). lactic acidemia. He was well until 19 months of age, when he experienced an acute febrile illness with anorexia, vomiting, and diarrhea. This was complicated by acute truncal ataxia, lethargy, and hypertonicity. | | Mutations: | A467T, PNF, PNF | | Age group: | infantile | | Age of Onset: 1.58, Age of Patient: n/a, Age of Death: 3.5 |
Back to top | Reference: | Martikainen et al, 2016; | | Description: | Parkinsons, PEO, short-term memory problems, general cerebral and cerebellar atrophy. | | Mutations: | A467T | | SNPs: | PNF | | Age group: | adult | | Age of Onset: 50, Age of Patient: 81, Age of Death: n/a |
| Reference: | Flemming, et al 2002; | | Description: | mental and motor developmental delay. At the age of 7 years, strabismus convergens\\ occurred. headache and nausea and\\ then presented with a generalized tonic-clonic seizure. myoclonia, focal motor seizures evolved into epilepsia partialis continua. Alpers. During the first 4 months of his illness, the boy became blind and incontinent and lost his ability to walk freely. During the\\ ensuing years, a further slight loss of mental and motor capabilities and several episodes of status epilepticus occurred. | | Mutations: | A467T | | SNPs: | PNF | | Age group: | childhood | | Age of Onset: 4, Age of Patient: 11, Age of Death: n/a |
| Reference: | Ashley et al, 2008; | | Description: | reported as Alpers, onset at 7 months, presenting encephalopathy with epilepsy, hepatopathy, and movement disorder (ataxia). 39% mtDNA copy number in muscle. | | Mutations: | A467T, T914P | | Age group: | infantile | | Age of Onset: 0.58, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Blok et al, 2009; | | Description: | Epilepsy, myoclonus, and developmental delay. | | Mutations: | A467T, T914P | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 1, Age of Death: n/a |
Back to top | Reference: | Horvath et al, 2006; | | Description: | Onset at 1.5 years with encephalopathy and hepatopathy and cortical blindness. Diagnosed as Alpers. | | Mutations: | A467T, T914P | | Age group: | infantile | | Age of Onset: 1.5, Age of Patient: 2, Age of Death: n/a |
| Reference: | Horvath et al, 2006; | | Description: | Onset at 4 years with encephalopathy, myoclonus, and SLE. | | Mutations: | A467T, T914P | | Age group: | childhood | | Age of Onset: 4, Age of Patient: 8, Age of Death: n/a |
| Reference: | Taanman et al, 2009; | | Description: | Onset at 9 months with alpers, death at 12 months. 8% mtDNA copy number in liver, 8% mtDNA copy number in muscle. | | Mutations: | A467T, T914P | | Age group: | infantile | | Age of Onset: 0.8, Age of Patient: n/a, Age of Death: 1 |
| Reference: | Tang et al, 2011; | | Description: | Developmental delay, headaches/migraines, seizures, hemiparesis, intractable seizure, high CSF lactate, high CSF protein, hearing loss, abnormal EEG, increased signal basal ganglia, abnormal MRI. 88% mtDNA copy number in blood. | | Mutations: | A467T, T914P | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 5, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Developmental delay, hypotonia, dementia/encephalopathy, seizures, intractable seizure, lactic acidosis, high CSF lactate, abnormal MRI, no liver problem at 3.5 years. 24% mtDNA copy number in blood. | | Mutations: | A467T, T914P | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 3, Age of Death: n/a |
Back to top | Reference: | Tang et al, 2011; | | Description: | Developmental delay, hypotonia, seizures, myoclonic seizures, hemiparesis, intractable seizure, elevated transaminases, FTT, lactic acidosis, high CSF lactate, elevated pyruvate, high CSF protein, dementia/encephalopathy. 112% mtDNA copy number in muscle, 63% mtDNA copy number in blood. | | Mutations: | A467T, T914P | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 0.9, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Developmental delay, seizures, myoclonic seizures, intractable seizure, respiratory deficiency/failure, abnormal EEG, abnormal MRI. 60% mtDNA copy number in blood. | | Mutations: | A467T, T914P | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 0.8, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Seizures, hepatic failure, duplicated cyst of ileum, FTT, diarrhea. 13% mtDNA copy number in liver. | | Mutations: | A467T, T914P | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 4, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Developmental delay, dementia, seizures, Alpers | | Mutations: | A467T, T914P | | Age group: | infantile | | Age of Onset: 2, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Onset 2 years with dementia, seizures, and liver failure. Alpers | | Mutations: | A467T, T914P | | Age group: | infantile | | Age of Onset: 2, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Dhamija et al, 2011; | | Description: | myoclonic status epilepticus, myoclonic seizures at 8 months, developmental regression, febrile upper respiratory tract infection, nearly continuous myoclonic jerks, normocephalic, nondysmorphic, diffuse hypotonia, died secondary to complications of hepatic failure. | | Mutations: | A467T, T914P | | Age group: | infantile | | Age of Onset: 0.8, Age of Patient: n/a, Age of Death: 1 |
| Reference: | Tzoulis et al, 2010; | | Description: | Encephalopathy with epilepsy, and liver disease. | | Mutations: | A467T, G303R | | Age group: | infantile | | Age of Onset: 0.9, Age of Patient: n/a, Age of Death: 1.1 |
Back to top | Reference: | Tzoulis et al, 2010; | | Description: | Encephalopathy with epilepsy, and liver disease. | | Mutations: | A467T, G303R | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: 1.4 |
| Reference: | Tzoulis et al, 2010; | | Description: | Encephalopathy with epilepsy, ataxia. | | Mutations: | A467T, G303R | | Age group: | infantile | | Age of Onset: 2, Age of Patient: n/a, Age of Death: 8 |
| Reference: | Sofou et al, 2012; | | Description: | Psychomotor deterioration, hypotonia, spasticity, Stroke-like episodes, Refractory seizures, Epilepsia Partialis Continua, Status Epilepticus, Myoclonus, severe hepatic dysfunction, | | Mutations: | A467T, G303R | | Age group: | infantile | | Age of Onset: 0.916, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Tzoulis et al, 2014; | | Description: | Epilepsy, stroke-like episode, Ataxia. | | Mutations: | A467T, G303R | | Age group: | infantile | | Age of Onset: 2, Age of Patient: n/a, Age of Death: 8 |
| Reference: | Tzoulis et al, 2014; | | Description: | Epilepsy, stroke-like episode. | | Mutations: | A467T, G303R | | Age group: | infantile | | Age of Onset: 0.9, Age of Patient: n/a, Age of Death: 1.1 |
Back to top | Reference: | Ashley et al, 2008; | | Description: | reported as Alpers, onset at 2 years, presenting encephalopathy with epilepsy, hepatopathy. 22% mtDNA copy number in muscle. | | Mutations: | A467T, R852C | | SNPs: | G11D | | Age group: | infantile | | Age of Onset: 2.25, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Stewart et al, 2009; | | Description: | Alpers, seizures, abnormal liver function, mild lactic acidosis, normal histology of COX fibers. | | Mutations: | A467T, R852C | | SNPs: | G11D | | Age group: | infantile | | Age of Onset: 1.25, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Seizures, myoclonic seizures, intractable seizures, hepatic failure, elevated transaminases, Apnes/Hypoventilation, respiratory deficiency/failure, lactic acidosis, organic aciduria/tiglyglycine, low plasma carnitine, abnormal EEG. 108% mtDNA copy number in blood. | | Mutations: | A467T, R852C | | SNPs: | G11D | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 0.8, Age of Death: n/a |
| Reference: | Hunter et al, 2011; | | Description: | Myoclonic Seizures, visual disturbance, motor paresis, Focal motor seizures, Epilepsia partialis continua, Developmental Delay or Regression, ataxia, Serum Lactate, liver mtDNA depletion, Alpers | | Mutations: | A467T, R852C | | Age group: | infantile | | Age of Onset: 1.33, Age of Patient: n/a, Age of Death: 5.08 |
Back to top | Reference: | McCoy et al, 2011; | | Description: | clonic seizures of the right arm and leg, Coma persisted and was accompanied by hypoglycaemia, tachycardia and oliguria, CSF proteinwas elevated, cortical blindness, anarthria and profound hypotonia, Liver dysfunction, prolonged focal seizures, acute encephalopathy, | | Mutations: | A467T, R852C | | Age group: | infantile | | Age of Onset: 1.75, Age of Patient: n/a, Age of Death: 3.5 |
| Reference: | Luoma et al, 2005; | | Description: | This 50-year-old woman developed progressive blepharoptosis slowly at the age of 40. She underwent blepharoplasty at the age of 47. Her neurological examination at the age of 50 was normal. Late onset ptosis | | Mutations: | A467T | | Age group: | adult | | Age of Onset: 40, Age of Patient: 50, Age of Death: n/a |
| Reference: | Luoma et al, 2005; | | Description: | This 64-year-old woman, the mother of the index patient, developed slowly progressive blepharoptosis at the age of 45. She underwent blepharoplasty at the age of 58. In the neurological examination at the age of 63, she had ptosis, but no other neurological deficits. Late onset ptosis. | | Mutations: | A467T | | Age group: | adult | | Age of Onset: 45, Age of Patient: 64, Age of Death: n/a |
| Reference: | Luoma et al, 2005; | | Description: | This 66-year-old woman developed progressive blepharoptosis slowly at the age of 50. She underwent blepharoplasty at the age of 62, and in the same year, she suffered a transitory ischemic attack. At the age of 66, no clinical findings except ptosis were detectable. Late onset ptosis. | | Mutations: | A467T | | Age group: | adult | | Age of Onset: 50, Age of Patient: 66, Age of Death: n/a |
| Reference: | Luoma et al, 2005; | | Description: | probable mild blepharoptosis, early onset ptosis | | Mutations: | A467T | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 23, Age of Death: n/a |
Back to top | Reference: | Galassi et al, 2008; | | Description: | at age 24 developed PEO and weakness of neck and proximal limb muscles. presence of multiple mtDNA deleted species, unsteady, broad-based gait. At age 32 she had complete PEO, bradykinesia, cogwheel rigidity, slurred speech, dysphagia, dysmetria, positive Romberg sign, reduced arm swing, and intermittent bilateral postural-action tremor. axonal sensorimotor neuropathy, severe depression, insomnia, and nocturnal panic attacks Has also mutation ANT1 gene (V289M) | | Mutations: | A467T | | Age group: | adult | | Age of Onset: 24, Age of Patient: 42, Age of Death: n/a |
| Reference: | Rouzier et al, 2013; | | Description: | LS-like, sensorimotor neuropathy. At 56 years old, patient 13 developed a motor neuron disease with severe tetraparesis, dysphonia and respiratory insufficiency leading to death a few years later. Muscular biopsy was strongly evocative of mitochondrial disease with a generalized RC deficiency, multiple mtDNA deletions and decreased mtDNA copy number in the muscle (39%). | | Mutations: | A467T | | Age group: | adult | | Age of Onset: 56, Age of Patient: 56, Age of Death: 60 |
| Reference: | Van Goethem et al, 2003a; | | Description: | sensory ataxic neuropathy, PEO, dysarthria. | | Mutations: | A467T, L304R | | Age group: | adult | | Age of Onset: 25, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Van Goethem et al, 2001; | | Description: | ocular symptoms, psychiatric symptoms, ptosis, opthalmoplegia, muscle weakness, distal sensory neuropathy, areflexia, pes cavus. | | Mutations: | A467T, L304R | | Age group: | adult | | Age of Onset: 25, Age of Patient: 38, Age of Death: n/a |
Back to top | Reference: | Van Goethem et al, 2001; | | Description: | ocular symptoms, ptosis, opthalmoplegia, dysphagia, muscle weakness, areflexia, retinopathy. | | Mutations: | A467T, L304R | | Age group: | adult | | Age of Onset: 24, Age of Patient: 34, Age of Death: n/a |
| Reference: | Van Goethem et al, 2001; | | Description: | psychiatric symptoms, ptosis, opthalmoplegia, dysphonia, dysphagia, muscle weakness, loss of weight, areflexia. | | Mutations: | A467T, L304R | | Age group: | juvenile | | Age of Onset: 16, Age of Patient: 28, Age of Death: n/a |
| Reference: | Ashley et al, 2008; | | Description: | Reported as Alpers, onset at 4 years, presenting encephalopathy with epilepsy. | | Mutations: | A467T, L966R | | Age group: | childhood | | Age of Onset: 4, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Hunter et al, 2011; | | Description: | Focal motor seizures, Epilepsia partialis continua, Developmental Delay or Regression, motor paresis, tremor Abnormal Liver Enzymes, Alpers | | Mutations: | A467T, L966R | | Age group: | infantile | | Age of Onset: 1.42, Age of Patient: n/a, Age of Death: 1.58 |
Back to top | Reference: | Hunter et al, 2011; | | Description: | Generalized tonic-clonic seizures, Focal motor seizures, Epilepsia partialis continua, Developmental Delay or Regression, visual disturbance, Abnormal Liver Enzymes, liver mtDNA depletion, Alpers | | Mutations: | A467T, L966R | | Age group: | infantile | | Age of Onset: 0.75, Age of Patient: n/a, Age of Death: 1.75 |
| Reference: | McCoy et al, 2011; | | Description: | focal seizures, elevated blood lactate, elevated liver enzymes, Elevated CSF protein, severe lactic acidosis, severe encephalopathy, Death occurred ten days after admission | | Mutations: | A467T, L966R | | Age group: | infantile | | Age of Onset: 1.42, Age of Patient: n/a, Age of Death: 1.42 |
| Reference: | Blok et al, 2009; | | Description: | Exercise intolerance, CPEO, retinitis pigmentosa, diabetes, limb girdle weakness. 2nd patient with cataract and myopathy | | Mutations: | A467T, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 51, Age of Death: n/a |
| Reference: | Stewart et al, 2009; | | Description: | PEO, ptosis, proximal weakness, multiple mtDNA deletions-LPCR. 25% COX-deficient fibers | | Mutations: | A467T, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 45, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Lovan et al, 2013; | | Description: | diabetes mellitus 2, congestive heart failure, hypothyroidism, hypertension, depression with psychotic features and gastric bypass surgery. gait difficulties, cognitive impairment, ophthalmoplegia, resting tremor and peripheral neuropathy. Shooting pains and numbness in her lower extremities, occasional rectal incontinence. Presyncopal dizziness. Complicated diabetic retinopathy. sensory ataxia secondary to a sensorimotor polyneuropathy, chronic supranuclear ophthalmoplegia, and a resting tremor of her right hand. bilateral ptosis and marked temporal muscle wasting. Her sensory examination found loss of pinprick sensation, temperature and vibration bilaterally up to her hips. She had bilateral vibration loss at her fingertips. She had loss of proprioception at her toes. She had bilateral dysdiadochokinesia with significant ataxia on finger to nose testing. Arreflexic in her lower extremities. On cognitive testing she was found to have moderate dementia with significant deficits in registration and construction as well as ideomotor apraxia. | | Mutations: | A467T, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: 55, Age of Patient: 60, Age of Death: n/a |
Back to top | Reference: | Blok et al, 2009; | | Description: | Alpers, died at age four. Sister also died at age 4 . | | Mutations: | A467T, S305R | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: n/a, Age of Death: 4 |
| Reference: | Tang et al, 2011; | | Description: | Liver failure. 51% mtDNA copy number in muscle, 9% mtDNA copy number in liver, 87% mtDNA copy number in blood. | | Mutations: | A467T, S305R | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 1, Age of Death: n/a |
| Reference: | Hunter et al, 2011; | | Description: | Myoclonic Seizures, Focal motor seizures, Developmental Delay or Regression, visual disturbance, Multi-organ failure, pancreatitis Abnormal Liver Enzymes, Serum Lactate, liver mtDNA depletion, Alpers | | Mutations: | A467T, S305R | | Age group: | childhood | | Age of Onset: 3, Age of Patient: n/a, Age of Death: 3.67 |
| Reference: | Blok et al, 2009; | | Description: | Epilepsy, liver failure, occipital strokes, and growth retardation, death at age 1. | | Mutations: | A467T, A957P | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: n/a, Age of Death: 1 |
| Reference: | de Vries et al, 2007; | | Description: | Onset at 8 months with FTT, dementia, hypotonia, seizures, hepatopathy, aplasia cutis, delayed myelinisation. Death at 17 months. | | Mutations: | A467T, A957P | | Age group: | infantile | | Age of Onset: 0.6, Age of Patient: n/a, Age of Death: 1.5 |
Back to top | Reference: | Ferrari et al, 2005; | | Description: | mild motor retardation was first noted at 6 months. At the age of 16 months, he was admitted to a local hospital for status epilepticus. At 18 months, generalized brain atrophy. epilepsia partialis continua, which was partially controlled by carbamazepine, later switched to oxcarbazepine. Elevated blood lactate. Alpers. He died at 2 years of age after severe complications from liver failure. | | Mutations: | A467T, A957P | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: n/a, Age of Death: 2 |
| Reference: | Horvath et al, 2006; | | Description: | Onset at 32 years with encephalopathy, PEO, ataxia, dysphagia, myopathy, neuropathy and cardiomyopathy, hearing loss. Death at 41 years. | | Mutations: | A467T, R627W | | Age group: | adult | | Age of Onset: 32, Age of Patient: n/a, Age of Death: 41 |
| Reference: | Horvath et al, 2006; | | Description: | Onset at 39 years with PEO, ataxia, myopathy, and hearing loss. | | Mutations: | A467T, R627W | | Age group: | adult | | Age of Onset: 39, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Horvath et al, 2006; | | Description: | PEO, SANDO (Horvath 2006 or Van goethem 2003) sensory ataxic neuropathy, PEO, dysarthria. | | Mutations: | A467T, R627W | | Age group: | adult | | Age of Onset: 20, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Kollberg et al, 2006; | | Description: | The girl was healthy and developed normally until one year of age when, during a mild respiratory infection, she had repeated multifocal partial seizures with clonic jerking and unconsciousness. The episode was later followed by the development of muscular hypotonia and of myoclonus. Mild psychomotor regression, ataxia, and slightly elevated serum transaminases were noticed. At 3 years of age, a mild infection provoked a prolonged status epilepticus, which lasted 3 weeks and included stroke-like features and rightsided hemiparesis. This was followed by a progressive deterioration of cognitive and motor functions and the development of ptosis and external ophthalmoplegia. Alpers. Febrile infections provoked repetitive status epilepticus with seizures in the form of migrating epilepsia partialis continua. mtDNA Deletions. | | Mutations: | A467T, R574W | | Age group: | infantile | | Age of Onset: 1, Age of Patient: 1.5, Age of Death: 4.333 |
Back to top | Reference: | Kollberg et al, 2006; | | Description: | mtDNA Deletions. She was healthy until 7 months of age when she, in association with pyelonephritis, had onset of myoclonic seizures and weakness of the left arm and leg. leftsided myoclonic seizures progressing to status epilepticus in the form of migrating EPC and unconsciousness. pneumonia and colitis. | | Mutations: | A467T, R574W | | Age group: | infantile | | Age of Onset: 0.583, Age of Patient: 1, Age of Death: 1.166 |
| Reference: | Spinazzola et al, 2009; | | Description: | Alpers. Alive at 10 years. Sister died at age 27. | | Mutations: | A467T, R574W | | Age group: | childhood | | Age of Onset: 3, Age of Patient: 10, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Authors report N/A. 6% mtDNA copy number in blood. | | Mutations: | A467T, F749S | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 0.8, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Intractable seizure, abrubt onset of seizure. 15% mtDNA copy number in blood. | | Mutations: | A467T, F749S | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 6, Age of Death: n/a |
Back to top | Reference: | Blok et al, 2009; | | Description: | FTT, died after epilepticus. | | Mutations: | A467T, R227P | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: n/a, Age of Death: 1 |
| Reference: | de Vries et al, 2007; | | Description: | Severe childhood multi-system disorder, epilepsy and failure to thrive, GI problems, hypotonia, retardation. | | Mutations: | A467T, R227P | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: n/a, Age of Death: 1 |
| Reference: | Ashley et al, 2008; | | Description: | Onset at 3 years of age, encephalopathy presenting with epilepsy and movement disorder (ataxia). | | Mutations: | A467T, C418R | | Age group: | childhood | | Age of Onset: 3, Age of Patient: n/a, Age of Death: n/a |
| Reference: | McCoy et al, 2011; | | Description: | presented in status epilepticus, with a 24 h history of lethargy and vomiting, ataxia diagnosed at 17 months, diffuse encephalopathy, | | Mutations: | A467T, C418R | | Age group: | infantile | | Age of Onset: 1.42, Age of Patient: n/a, Age of Death: 3.58 |
| Reference: | Kurt et al, 2010; | | Description: | Psychomotor delay, seizures, liver disease, GI dysmotility, lactic acidosis, ptosis, hearing loss. | | Mutations: | A467T, P1073L | | Age group: | infantile | | Age of Onset: 0.01, Age of Patient: n/a, Age of Death: 0.8 |
Back to top | Reference: | Kurt et al, 2010; | | Description: | Psychomotor delay, status epilepticus, liver disease, GI dysmotility, lactic acidosis, optic atrophy. | | Mutations: | A467T, P1073L | | Age group: | infantile | | Age of Onset: 0.01, Age of Patient: n/a, Age of Death: 3 |
| Reference: | Tang et al, 2011; | | Description: | Developmental delay, seizures, abnormal MRI, hypotonia, low in blood. | | Mutations: | A467T, C1188R | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 3, Age of Death: n/a |
| Reference: | Khan et al, 2012; | | Description: | developmental delay, prolonged generalized seizure, hypotonia, epilepsia partialis continua, congestive heart failure, respiratory difficulty, autopsy showed enlarged and yellow liver, symmetrical but multifocal ischemic encephalopathy with laminar necrosis and neuronal degeneration in the cerebral cortex, Mitochondrial DNA copy number in blood was 43% of control values | | Mutations: | A467T, C1188R | | Age group: | infantile | | Age of Onset: 0.75, Age of Patient: n/a, Age of Death: 1.17 |
| Reference: | Wong et al, 2008; | | Description: | Onset 60 years with ataxia, neuropathy, myopathy, hearing loss, cerebellar atrophy, SANDO/SCAE, PEO. | | Mutations: | A467T, G737R | | Age group: | adult | | Age of Onset: 60, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Milone et al, 2011; | | Description: | progressive sensory ataxic neuropathy, ophthalmoparesis, cerebellar ataxia, limb weakness, muscle cramps, sensory hearing loss, dysarthria, dysphagia, constipation, and memory loss, brain MRI was abnormal with evidence of generalized cortical and cerebellar atrophy, evidence of a length-dependent sensory > motor polyneuropathy of axonal type, Multiple mtDNA deletions detected by PCR in blood | | Mutations: | A467T, G737R | | Age group: | adult | | Age of Onset: 53, Age of Patient: 58, Age of Death: n/a |
Back to top | Reference: | Wong et al, 2008; | | Description: | Onset 48 years with neuropathy, myopathy, SANDO, lactic acidosis PEO. | | Mutations: | A467T, R1138C | | Age group: | adult | | Age of Onset: 48, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Milone et al, 2011; | | Description: | Bilateral ptosis, distal lower limbs paresthesias, dysphagia, imbalance, inattention, ophthalmoparesis, Multiple mtDNA deletions detected by PCR in blood and muscle | | Mutations: | A467T, R1138C | | Age group: | adult | | Age of Onset: 46, Age of Patient: 54, Age of Death: n/a |
| Reference: | Luoma et al, 2005; | | Description: | developed progressive blepharoptosis at the age of 20 and ophthalmoplegia during subsequent years, at 35, the patient developed progressive unsteadiness of gait. She noted fatigue, myalgia and cramps, which were triggered by moderate exercise. At 44, restless legs syndrome, deficits in memory and concentration, and suffered from recurrent episodes of depression, at 46 showed bilateral blepharoptosis, limited eye movements in all directions except downwards, lateral rotatory nystagmus and slight upbeat nystagmus in upward gaze, but no saccades. She had facial muscle weakness, dysarthria and slight proximal muscle weakness. She had glove and stockinglike hypoesthesia, distally impaired vibration sense, absent ankle reflexes and weak other tendon reflexes, as well as mild dysmetria. Her gait was broad-based, with further impairment with eyes closed. Tandem gait was impaired. deltoid muscle showed myopathic features, structurally abnormal mitochondria with para-crystalline inclusions, | | Mutations: | A467T, R627Q | | SNPs: | Q1236H | | Age group: | adult | | Age of Onset: 20, Age of Patient: 46, Age of Death: n/a |
| Reference: | Hanisch et al, 2014; | | Description: | Ataxia, ptosis, myalgia, sensory neuropathy, motor neuropathy, axonal neuropathy, demyelinating neuropathy. | | Mutations: | A467T, R627Q | | Age group: | adult | | Age of Onset: 46, Age of Patient: 50, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Alpers, chorea, microcephaly, leukodystrophy, Seizures, Dementia, developmental delay. | | Mutations: | A467T | | SNPs: | PNF, Q1236H | | Age group: | infantile | | Age of Onset: 1.5, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Hunter et al, 2011; | | Description: | Developmental Delay or Regression, hypotonia, vomiting, Abnormal Liver Enzymes, liver mtDNA depletion, clinical diagnosis of infantile hepatopathy | | Mutations: | A467T, H277L, R232H | | Age group: | infantile | | Age of Onset: 0.125, Age of Patient: n/a, Age of Death: 0.25 |
| Reference: | Hunter et al, 2011; | | Description: | Developmental Delay or Regression, motor paresis, hypotonia, vomiting, Abnormal Liver Enzymes, Serum Lactate, liver mtDNA depletion, clinical diagnosis of infantile hepatopathy | | Mutations: | A467T, H277L, R232H | | Age group: | infantile | | Age of Onset: 0.17, Age of Patient: n/a, Age of Death: 0.25 |
| Reference: | Van Goethem et al, 2001; | | Description: | ptosis, opthalmoplegia, dysphonia, dysphagia, distal sensory neuropathy, areflexia. | | Mutations: | A467T | | SNPs: | R3P | | Age group: | adult | | Age of Onset: 30, Age of Patient: 58, Age of Death: n/a |
| Reference: | Van Goethem et al, 2001; | | Description: | ptosis, opthalmoplegia, dysphonia, dysphagia, muscle weakness, loss of weight, distal sensory neuropathy, areflexia. | | Mutations: | A467T | | SNPs: | R3P | | Age group: | adult | | Age of Onset: 20, Age of Patient: 61, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Clinical indications reported as N/A only information provided is age of patient and 71% mtDNA copy number in blood. | | Mutations: | A467T, F88L | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 42, Age of Death: n/a |
Back to top | Reference: | Tang et al, 2011; | | Description: | Dementia/encephalopathy, headaches/migraines, ataxia, seizures, peripheral neuropathy, exercise intolerance, muscle weakness, easy fatigability, CPEO, abnormal EMG/NCV, hearing loss, cerebellar atrophy, abnormal MRI. 82% mtDNA copy number in blood. | | Mutations: | A143V, A467T | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 49, Age of Death: n/a |
| Reference: | de Vries et al, 2007; | | Description: | Onset at 4 months with FTT, dementia, hypotonia, seizures, myoclonus, GI problems, hepatopathy, hearing loss, delayed myelinisation. Death at 10 months | | Mutations: | A467T, R227W | | Age group: | infantile | | Age of Onset: 0.3, Age of Patient: n/a, Age of Death: 0.8 |
| Reference: | Ashley et al, 2008; | | Description: | onset at 2 years with encephalopathy no hepatopathy. | | Mutations: | A467T, R417T | | Age group: | infantile | | Age of Onset: 2, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Sarzi et al, 2007; | | Description: | Alpers, cerebellar ataxia at age 2, seizures at age 3, RC deficiency in liver, hepatocellular insufficiency after valproate treatment at age 3.6, death at age 3.8. 8% mtDNA copy number in liver. | | Mutations: | A467T, L428P | | Age group: | infantile | | Age of Onset: 2, Age of Patient: n/a, Age of Death: 3.8 |
| Reference: | Tang et al, 2011; | | Description: | Hypotonia, hepatic failure, elevated transaminases, respiratory deficiency/failure, FTT, developmental delay, GI reflux, delayed gastric emptying, dicarboxylic aciduria, high CSF protein, abnormal muscle histology, COX deficiency, vomiting. 12% mtDNA copy number in muscle, 42% mtDNA copy number in blood, ETC low. ptosis, poor head control. Central hypotonia, and absent deep tendon reflexes. He developed hypoxia, ascites, tachycardia, intermittent fevers, and sepsis. Some severely cytochrome oxidase-deficient muscle fibers were present. | | Mutations: | A467T, S1095R | | Age group: | infantile | | Age of Onset: 0.333, Age of Patient: n/a, Age of Death: 0.583 |
Back to top | Reference: | Tang et al, 2011; | | Description: | Encephalopathy/dementia, liver failure, cholestasis, lactic acidosis, altered mental status. 47% mtDNA copy number in muscle, 8% mtDNA copy number in liver, ETC low. | | Mutations: | A467T, R807C | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 1, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Developmental delay, hypotonia, seizures, hepatic failure, elevated transaminases, renal tubular disease, failure to thrive. 18% mtDNA copy number in blood. | | Mutations: | A467T, R807H | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 1, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Onset 1 years with dementia, seizures, and liver failure. Alpers | | Mutations: | A467T, L886P | | Age group: | infantile | | Age of Onset: 1, Age of Patient: 1, Age of Death: n/a |
| Reference: | Horvath et al, 2006; | | Description: | Onset at .5 years with encephalopathy, liver dysfunction, cardiopathy, diagnosed as Alpers, death at 1.3 years. | | Mutations: | A467T, K1191N | | Age group: | infantile | | Age of Onset: 0.5, Age of Patient: n/a, Age of Death: 1.3 |
| Reference: | Stewart et al, 2009; | | Description: | Alpers, seizures, myoclonus, liver failure, elevated serum lactate, COX deficient fibers present in muscle and liver, mtDNA depletion in liver. | | Mutations: | A467T, L605R | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Tang et al, 2011; | | Description: | VPA induced liver failure. 50% mtDNA copy number in muscle. | | Mutations: | A467T, G588D | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 2, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Developmental delay, intractable seizure/refractory, hepatic failure, cerebellar atrophy, liver failure. 87% mtDNA copy number in blood. | | Mutations: | A467T, H754Q | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 2, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Dementia/encephalopathy, ataxia, peripheral neuropathy, ptosis, abnormal muscle histology, abnormal muscle ultrastructure, abnormal respiratory enzyms, large mitochondrial/ proliferation, COX deficiency, ragged red fibers. 100% mtDNA copy number in blood. | | Mutations: | A467T, R597W | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 26, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Encephalopathy, seizures, pancreatitis, hepatic failure, elevated transaminases, respiratory deficiency/failure, lactic acidosis, mycoplasma infection, pentobarbital induced liver failure. 57% mtDNA copy number in muscle, 78% mtDNA copy number in blood, Complex IV 20%. | | Mutations: | A467T, L1113P | | Age group: | infantile | | Age of Onset: n/a, Age of Patient: 1, Age of Death: n/a |
| Reference: | Baruffini et al, 2011; | | Description: | Seizures onset at age 1, death via VPA induced liver failure at age 2. | | Mutations: | A467T, P625R | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: 2 |
Back to top | Reference: | Agostino et al, 2003; | | Description: | PEO, bilateral ptosis, severe limita- tion of ocular motility, and a mosaic distribution of ragged-red and cytochrome c oxidase–negative fibers in the muscle biopsy. All patients had multiple deletions of muscle mtDNA. | | Mutations: | A467T, S1104C | | Age group: | adult | | Age of Onset: 48, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Di Fonzo et al, 2003; | | Description: | PEO, age of onset 65 years. | | Mutations: | A467T, G268A | | Age group: | adult | | Age of Onset: 65, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Echaniz-Laguna et al, 2010; | | Description: | presented with blurred vision, elevated CSF protein, she successively developed various neurological signs, ie, bilateral external ophthalmoplegia, ataxia, hearing impairment, and severe depression, ptosis, myopathy, cardiomyopathy, and dysphagia, Muscle biopsy performed at 63 years of age showed numerous ragged-red fibers and multiple mtDNA deletions | | Mutations: | A467T, R275Q | | Age group: | adult | | Age of Onset: 30, Age of Patient: 63, Age of Death: n/a |
| Reference: | Lax et al, 2012a; | | Description: | CPEO, Ptosis, Peripheral neuropathy, COX-deficient fibers, ragged red fibers, presence of mitochondrial dna deletions in muscle, Sensory and motor neuronopathy, Distal and proximal neurogenic change | | Mutations: | A467T, R1096C | | Age group: | juvenile | | Age of Onset: 17, Age of Patient: 42, Age of Death: n/a |
| Reference: | Amiot et al, 2009; | | Description: | Peripheral neuropathy, PEO, ataxia | | Mutations: | A467T, M919T | | Age group: | adult | | Age of Onset: 23, Age of Patient: 41, Age of Death: n/a |
Back to top | Reference: | Sitarz et al, 2014; | | Description: | Epilepsy, Ataxia, Myopathy. | | Mutations: | A467T, S1104F | | Age group: | childhood | | Age of Onset: n/a, Age of Patient: 5, Age of Death: n/a |
| Reference: | McFarland et al, 2008; | | Description: | Presented with epilepsy following minor head trauma, 2 months after valproate treatment developed persistent vomiting and encephalopathy, deranged liver function tests, prolonged clotting, elevated ammonia, and a high plasma lactate, Hepatic dysfunction progressed, | | Mutations: | A467T, Q879H, T885S | | SNPs: | E1143G | | Age group: | infantile | | Age of Onset: 2, Age of Patient: n/a, Age of Death: n/a |
|
|
|
|
|
The following information is based on existing patient data and pathogenic cluster assignment.
Pathogenicity information for a patient with mutation A467T and a cluster P587L mutation: Age of onset information is extracted from a total of 3 patients and/ or patient families. | Age of onset | | |
3- 2- | 0
| 0
| 0
| 3
| | | infantile | childhd | juvenile | adult | | | 0% | 0% | 0% | 100% | |
All mutations mapping within the pathogenic clusters are at high risk for pathogenicity. In general, a patient must have a pathogenic mutation in both of his/ her POLG genes to develop a POLG-related syndrome.  | Symptoms described in patients with A467T-P587L mutations | | Symptoms in patients with combination
A467T:P587L | | Ptosis | 66.7% | | PEO | 66.7% | | Movement disorder (ataxia) | 33.3% | | Sensory ataxia | 33.3% | | Peripheral neuropathy | 33.3% | | Polyneuropathy | 33.3% | | Axonal sensorimotor polyneuropathy | 33.3% | | Exercise intolerance | 33.3% | | Myopathy | 33.3% | | Ophthalmoplegia | 33.3% | | Dementia | 33.3% | | Proximal weakness | 33.3% | | Hypothyroidism | 33.3% | | Tremor | 33.3% |
| | Data gathered from clinical descriptions for 3 patients |
| Symptoms by group | | CPEO | 100.0% | | Other | 66.7% | | Ataxia | 33.3% | | CNS symptoms | 33.3% | | Developmental Delay | 33.3% | | Myopathy | 33.3% | | Neuropathy | 33.3% |
| | [Show grouping information] |
|  | Symptoms described in patients with cluster2-cluster2 mutations | |
| Symptoms in patients with combination
cluster2:cluster2 | | Movement disorder (ataxia) | 53.6% | | PEO | 36.2% | | Ptosis | 27.5% | | Epilepsy | 26.1% | | Peripheral neuropathy | 26.1% | | Myoclonic seizures | 24.6% | | Status epilepticus | 18.8% | | Encephalopathy | 18.8% | | Ophthalmoplegia | 17.4% | | Dysarthria | 15.9% | | Nystagmus | 14.5% | | Ragged red fibers | 13.0% | | Myopathy | 13.0% | | Stroke | 13.0% | | Headache/ migraine | 13.0% | | Developmental delay | 13.0% | | Epilepsia partialis | 11.6% | | Demyelinating neuropathy | 11.6% | | Sensory ataxia | 10.1% | | Muscle weakness | 10.1% | | Liver failure | 8.7% | | Tremor | 8.7% | | Cox-negative | 7.2% | | Dysphagia | 7.2% | | +34 other symptoms in under 5.0% of the patients |
| | Data gathered from clinical descriptions for 69 patients |
| Symptoms by group | | CPEO | 58.0% | | Ataxia | 55.1% | | Seizures | 53.6% | | CNS symptoms | 43.5% | | Neuropathy | 40.6% | | Myopathy | 31.9% | | Developmental Delay | 23.2% | | Hepatopathy | 15.9% | | GI symptoms | 13.0% | | Migraines | 13.0% | | Other | 13.0% | | Alpers syndrome | 10.1% | | Hypotonia | 4.3% | | Unknown | 1.4% |
| | [Show grouping information] |
|
|
|